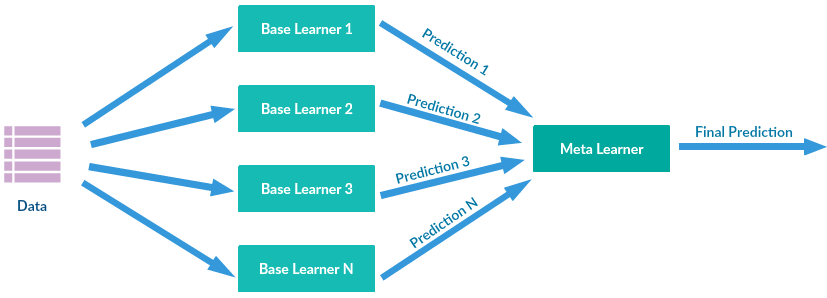
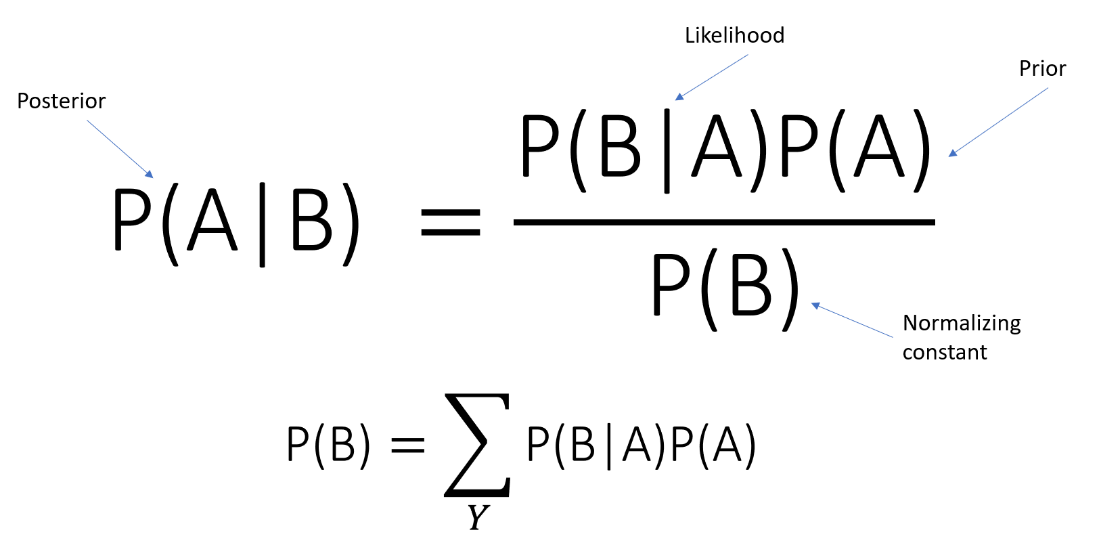This repository contains implementations of six Machine Learning Classifiers using only standard Python libraries such as NumPy and Pandas.
- compatible with python 3.6
The current repository includes 6 implementations of Models:
- Naïve Bayes Classifier at
./src/naive_bayes.py - Decision Tree at
./src/decision_tree.py - Logistic Regression at
./src/logistic_regression.py - Random Forest at
./src/random_forest.py - Adaboost at
./src/adaboost.py - Stacking at
./src/stacking.py
class NaiveBayes(type = “Gaussian”, prior = None)This class implements Naïve Bayes with Gaussian and Multinomial types.
| Parameters | type: str, ‘Gaussian’ or ‘Multinomial’, default: ‘Gaussian’ Specifies which type ‘Gaussian’ or ‘Multinomial’ is to be used. prior: array-like, shape (n_classes,) Prior probabilities of the classes. If specified the priors are not adjusted according to the data. |
| Attributes | class_log_prior: array, shape(n_classes) Prior probability of each class._ |
def fit(X, y)Fit the model according to the given training data.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y: array-like, shape = (n_samples,) Target vector relative to X. |
| Returns | self: Object of class NaiveBayes |
def predict(X)Get the class predictions.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
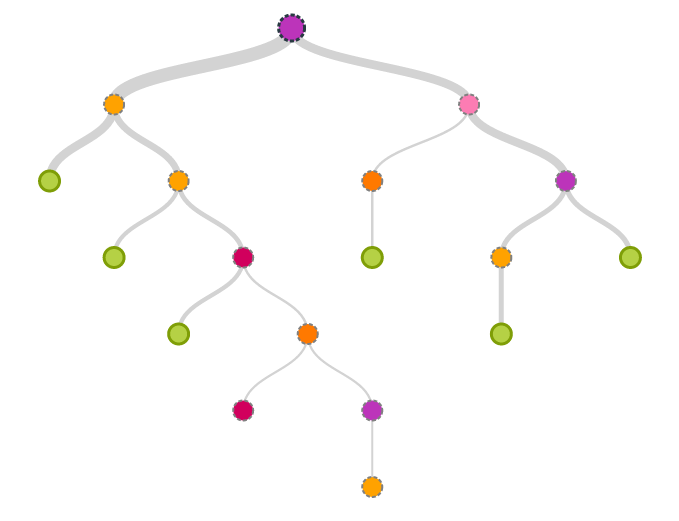
class DecisionTree(max_depth = 1,split_val_metric = 'mean', min_info_gain = 0.0,
split_node_criterion = 'gini')This class implements the Decision Tree Classifier.
| Parameters | max_depth: int, default: 1 The maximum depth upto which the tree is allowed to grow. split_val_metric: str, ‘mean’ or ‘median’, default: ‘mean’ This is the value of the column selected on which you make a binary split. min_info_gain: float, default: 0.0 Minimum value of information gain required for node to split. split_node_criterion: str, ‘gini’ or ‘entropy’, default: ‘gini’ Criterion used for splitting nodes. |
def build_tree(X, y, sample_weights)Build the Decision Tree according to the given data.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y: array-like, shape = (n_samples,) Target vector relative to X. sample_weight: array-like, shape = [n_samples] or None Sample weights. If None, then samples are equally weighted. Splits that would create child nodes with net zero or negative weight are ignored while searching for a split in each node. Splits are also ignored if they would result in any single class carrying a negative weight in either child node. |
| Returns | self: Object of class DecisionTree |
def predict_new(X)Get the class predictions.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
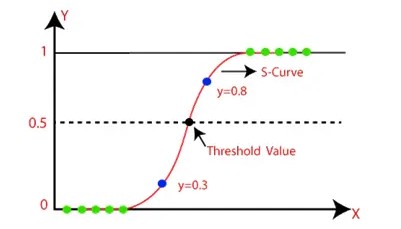
class LogisticRegression(regulariser = 'L2', lamda = 0.0, num_steps = 100000, learning_rate = 0.1,
initial_wts = None, verbose=False)This class implements the Logistic Regression Classifier with L1 and L2 regularisation.
| Parameters | regulariser: str, ‘L1’ or ‘L2’, default: ‘L2’ Specifies whether L1 or L2 norm is to be used for regularisation. lamda: float, default: 0.0 Hyperparameter for regulariser term. Default value indicates no regularisation is to be done. Directly proportional to regularisation strength. num_steps: int, default: 100000 Number of iterations for which Gradient Descent will run. learning_rate: float, default: 0.1 Indicates the size of step taken for Gradient Descent. initial_wts: array, shape(n_features+1,), default: None A list of n+1 values, where n is number of features. This is the initial value of the coefficients. If None, values of list assigned randomly in range normal (0,1). verbose: bool, default: False Whether to display loss after every 1000 iterations. |
| Attributes | theta_: array, shape(n_features,) A list of weights. bias_: int Bias term for Logistic Regression equation. para_theta_: dictionary, {(class, theta_)} A dictionary with the weights corresponding to each class. para_bias_: dictionary, {(class, bias_)} A dictionary with the biases corresponding to each class. hypothesis: dictionary, {(class, probs)} A dictionary with the probabilities of each sample corresponding to each class. |
def fit(X, y)Fit the model according to the given training data.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y: array-like, shape = (n_samples,) Target vector relative to X. |
| Returns | self: Object of class NaiveBayes |
def predict_prob(X)Returns probability estimates for all classes ordered by label of classes.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | T: array-like, shape = (n_samples, class) Returns the class for each sample according to whether n_classes>2 or not. |
def predict(X)Predicts the class labels for samples in X.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
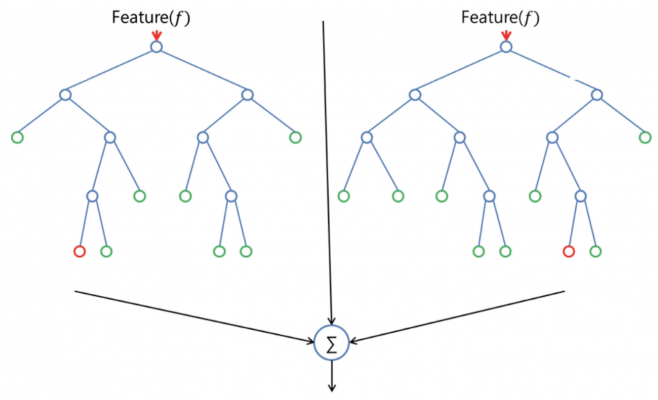
class RandomForest(n_trees=10, max_depth=5, split_val_metric='mean', min_info_gain=0.0,
split_node_criterion='gini', max_features=5, bootstrap=True, n_cores=1)This class implements the Random Forest Classifier.
| Parameters | n_trees: int, default: 10 Number of estimator trees to build the ensemble. max_depth: int, default: 1 The maximum depth upto which the tree is allowed to grow. split_val_metric: str, ‘mean’ or ‘median’, default: ‘mean’ This is the value of the column selected on which you make a binary split. min_info_gain: float, default: 0.0 Minimum value of information gain required for node to split. split_node_criterion: str, ‘gini’ or ‘entropy’, default: ‘gini’ Criterion used for splitting nodes. max_features: int, default: 5 The number of features to consider when looking for the best split. bootstrap: bool, default: True Whether bootstrap samples are to be used. If False, the whole dataset is used to build each tree. n_cores: int, default: 1 Number of cores on which to run this learning process. |
def fit_predict(X_train, y_train, X_test)Fits the model according to the given training data and predicts the class label for sample X_test.
| Parameters | X_train: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y_train: array-like, shape = (n_samples,) Target vector relative to X. X_test: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
class AdaboostClassifier(max_depth = 1, n_trees = 10, learning_rate = 0.1)This class implements Adaboost for Multiclass Classification.
| Parameters | max_depth: int, default: 1 The maximum depth upto which the tree is allowed to grow. n_trees: int, default: 10 Number of estimator trees to build the ensemble. learning_rate: float, default: 0.1 Used in calculating error weights. |
| Attributes | trees_: list, shape: (n_trees,) A list of tree estimators. tree_weights_: array, shape: (n_trees,) Weights of tree estimators. tree_errors_: array, shape: (n_trees,) Used to store errors for tree estimators. n_classes: int Number of disctinct classes. classes_: array, shape: (n_classes,) Array of class labels. n_samples: int Number of sample examples under consideration. |
def fit(X, y)Fit the model according to given training data.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y: array-like, shape = (n_samples,) Target vector relative to X. |
| Returns | self: Object of class AdaboostClassifier |
def predict(X)Predicts the class labels for samples in X.
| Parameters | X: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
class Stack([(clf, count)])This class implements the Stacking Ensemble Classifier.
| Parameters | [(clf, count)]: list of tuples First element of each tuple is a Classifier object. Second element of each tuple is how many times it is to be considered. |
def fit_pred(X_train, X_test, y_train)Fits the model according to the given training data and predicts the class label for sample X_test.
| Parameters | X_train: {array-like, sparse matrix}, shape = (n_samples, n_features) Training Vector where n_samples is the number of samples and n_features is the number of features. y_train: array-like, shape = (n_samples,) Target vector relative to X. X_test: {array-like, sparse matrix}, shape = (n_samples, n_features) Input Vector where n_samples is the number of samples and n_features is the number of features. |
| Returns | C: array-like, shape = (n_samples) Predicted class label per sample. |
For any clarification, comments, or suggestions please contact Arkil.