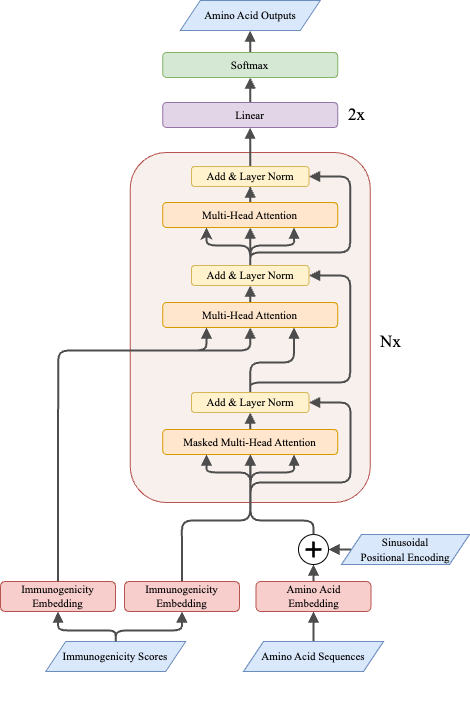
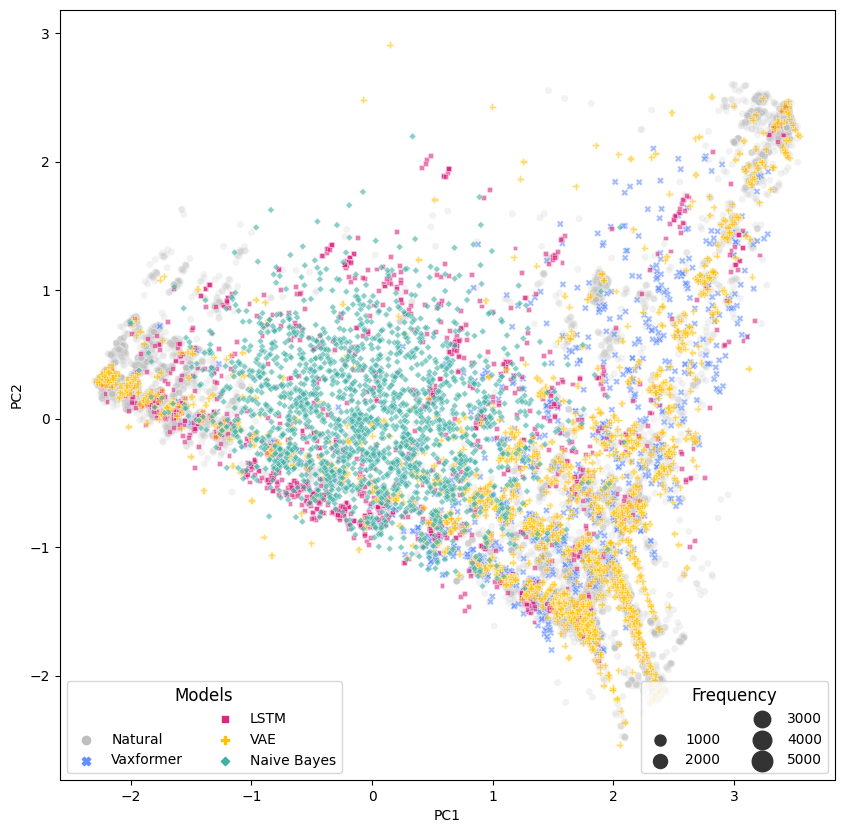
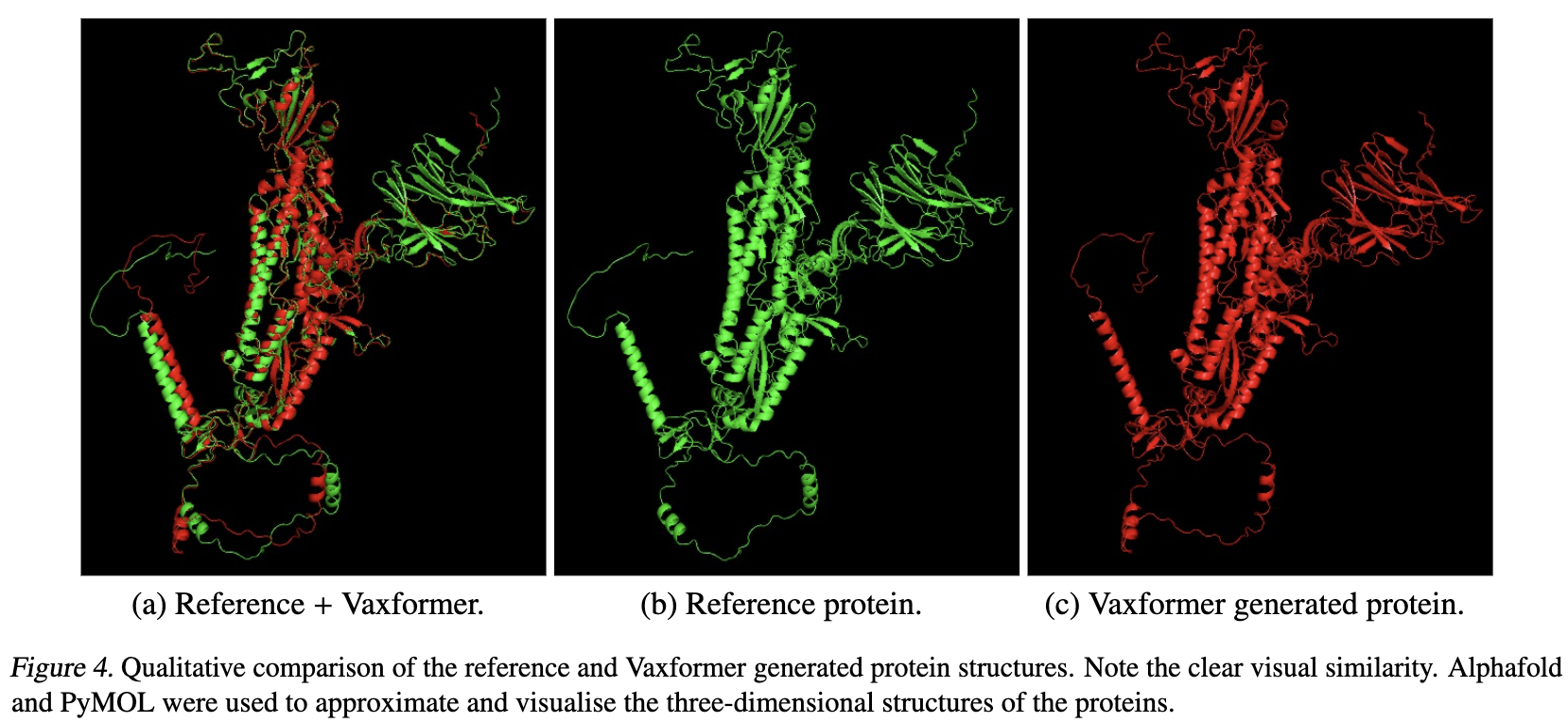
This repository contains pre-trained models, corpora, indices, and code for pre-training, finetuning, retrieving and evaluating for "Vaxformer: Antigenicity-controlled Transformer for Vaccine Design Against SARS-CoV-2"
The SARS-CoV-2 pandemic has emphasised the importance of developing a universal vaccine that can protect against current and future variants of the virus. However, research in the field of protein design is limited despite the advancements in AI and our knowledge of the human immune system. In this context, the present study proposes a novel conditional protein Language Model architecture, called Vaxformer, which is designed to produce antigenicity-controlled SARS-CoV-2 spike proteins. We evaluate the generated protein sequences of the Vaxformer model using DDGun protein stability measure, netMHCpan antigenicity score, and a structure fidelity score using the root mean square deviation between the protein folded with AlphaFold and a Covid reference protein to gauge its viability for vaccine development. Our results show that Vaxformer outperforms the existing state-of-the-art Conditional Variational Autoencoder model to generate antigenicity-controlled SARS-CoV-2 spike proteins. These findings suggest promising opportunities for Conditional Transformer models to expand our understanding of vaccine design and their role in mitigating global health challenges.
Authors (equal contribution):
This codebase requires the following dependencies:
- biopython
- matplotlib
- seaborn
- numpy
- pandas
- pydantic
- python-dotenv
- PyYAML
- tqdm
- wandb
- jupyterlab
We opted in to using conda as our package manager. The following will install all necessary dependencies for a GPU training:
ENV_NAME=vaxformer
conda create -n ${ENV_NAME} python=3.8 -y
conda activate ${ENV_NAME}
conda install pytorch torchvision torchaudio pytorch-cuda=11.7 -c pytorch -c nvidia -y
pip install -r requirements.txt
Note
netMHCpan only runs on Linux or Darwin machine
Follow this step to setup netMHCpan:
- Download https://services.healthtech.dtu.dk/services/NetMHCpan-4.1/
- Follow their installation instruction outlined in the
netMHCpan-4.1.readmefile
We included DDGun as a submodule to this repo. To install, you need to:
# Clone the DDGun submodule
git submodule update --init --recursive
# Install DDGun
cd submodules/ddgun
python setup.py
- Install PyMol
- Install alphafold2 or use HPC cluster with alphafold installed (recommended)
Vaxformer is trained with a dataset of spike Covid proteins from GI-SAID. You have to have the appropriate GI-SAID credentials to download the dataset. To obtain comparable data splits, we inquired to the author of the previous publication (ImmuneConstrainedVAE).
.
├── configs # Config files
│ ├── test/ # Config files for sampling runs
│ └── train/ # Config files for training runs
├── datasets/ # Datasets of sequences and immunogenicity scores
├── scripts # Scripts to start runs
│ ├── netmhcpan/
│ │ ├── netmhcpan_allele_scores_one_file.sh # Script to run netMHCpan scoring for multiple peptide files
│ │ ├── netmhcpan_allele_scores.sh # Script to run netMHCpan scoring for one peptide file
│ │ ├── compute_hits_from_peptides.py # Script to compute netMHCpan hits and scores from peptides
│ │ └── generate_peptides_from_sequences.py # Script to generate peptides from sequences
│ ├── evaluation/
│ │ ├── alphafold.sh # Script to run the AlphaFold evaluation
│ │ ├── ddGun.sh # Script to run the DDGun evaluation
│ │ ├── create_data_for_ddgun.ipynb # Notebook to create the necessary data for DDGun run
│ │ ├── dd_eval.ipynb # Notebook to read the DDGun evaluation and find the top 10 proteins
│ │ └── evaluation.ipynb # Notebook for PCA evaluation
│ ├── baseline/
│ │ └── naive_bayes.ipynb # Notebook for
│ ├── slurm/ # Slurm scripts for training and sampling runs
│ ├── sample.py # Script to run sampling with a model of choice
│ └── train.py # Script to run training with a model configuration of choice
├── src
│ ├── dataset
│ │ ├── __init__.py
│ │ ├── dataset.py # Dataset class to prepare and iterate through the dataset
│ │ └── tokenizer.py # Tokenizer class to preprocess the input sequence
│ ├── models
│ │ ├── __init__.py
│ │ ├── baseline.py # Naive Bayes baseline model
│ │ ├── lstm.py # Conditional LSTM model
│ │ ├── vae.py # Conditional VAE model
│ │ └── vaxformer.py # The proposed Vaxformer model
│ ├── utils
│ │ ├── __init__.py
│ │ ├── common_utils.py # Utility functions to prepare trainings
│ │ └── model_utils.py # Utility functions for modelling purposes
│ ├── __init__.py
│ ├── configs.py # Pydantic configs validator
│ ├── constants.py # Constants for the training
│ └── trainer.py # Trainer class to handle training operations
├── submodules
│ └── ddgun/ # The DDGun package that we use for one of the evaluations
├── requirements.txt # Necessary Python Packages
├── README.md # You are here
Once you obtained the dataset, you need to first run netMHCpan to all sequences to compute the immunogenicity scores. First, you need to generate 9-mer peptides of each sequence of each dataset split:
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/TRAIN_DATASET.txt
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/VALID_DATASET.txt
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/TEST_DATASET.txt
These runs will create .pep files containing 9-mer peptides for each sequences which then can be passed to the netMHCpan:
cd scripts/netmhcpan/
bash netmhcpan_allele_scores_one_file.sh PATH/TO/TRAIN_DATASET_PEPTIDES_FILE.pep
bash netmhcpan_allele_scores_one_file.sh PATH/TO/VALID_DATASET_PEPTIDES_FILE.pep
bash netmhcpan_allele_scores_one_file.sh PATH/TO/TEST_DATASET_PEPTIDES_FILE.pep
These runs will create .pep.out files which contains the immunogenicity score for each peptides. Finally, we need to reconcile the peptides into sequences and calculate the hits scores of each sequence:
python scripts/netmhcpan/compute_hits_from_peptides.py \
--sequences_filepath=PATH/TO/TRAIN_DATASET.txt \
--peptides_dir=PATH/TO/TRAIN_PEPTIDES_DIR/ \
--peptide_file_prefix=TRAIN_PEPTIDE_FILE_PREFIX
python scripts/netmhcpan/compute_hits_from_peptides.py \
--sequences_filepath=PATH/TO/VALID_DATASET.txt \
--peptides_dir=PATH/TO/VALID_PEPTIDES_DIR/ \
--peptide_file_prefix=VALID_PEPTIDE_FILE_PREFIX
python scripts/netmhcpan/compute_hits_from_peptides.py \
--sequences_filepath=PATH/TO/TEST_DATASET.txt \
--peptides_dir=PATH/TO/TEST_PEPTIDES_DIR/ \
--peptide_file_prefix=TEST_PEPTIDE_FILE_PREFIX
Each of the peptide files that are generated from the previous command would have a prefix. For instance, from the previous commands you obtained 12 files (vaxformer_large_A0101.pep.out, ... , vaxformer_large_B5801.pep.out), then the peptide file prefix is vaxformer_large.
Practically, we can obtain quantiles from the distribution of hits of the training data split. For the sake of simplicity and reproducibility (and sanity check), you can check the Q1 and Q3 that we used to calculate the immunogenicity scores in the src/constants.py which are denoted as IMMUNOGENICITY_Q1 and IMMUNOGENICITY_Q3 respectively.
These quantiles are used to compute the immunogenicity scores of each sequence. Sequences whose scores are lower than the Q1 are considered to have low immunogenicity scores (0), in between Q1 and Q3 are intermediate immunogenicity scores (1), and above Q3 are high immunogenicity scores (3).
Once the sequences and immunogenicity scores datasets are obtained, we can run a training process.
python scripts/train.py \
--config_filepath=PATH/TO/TRAIN_CONFIG_FILE
Selections of train config files can be found in the configs/train/ folder.
Both LSTM and Vaxformer can be trained with -small, -base, or -large setting. They differ in terms of the number of hidden layers and their sizes.
Before any evaluation steps, we need to first generate the sequences with a pretrained model of choice
python scripts/sample.py \
--config_filepath=PATH/TO/TEST_CONFIG_FILE
--num_sequences=2000
Examples of test config files can be found in the configs/test/ folder.
- Use create_data_for_ddgun.ipynb provided in scripts folder to generate input for ddgun
- Install DDGun (https://github.com/biofold/ddgun)
- Use ddGun.sh provided in scripts folder
- dd_eval.ipynb provides functions necessary to read the output of the DDGun
To evaluate the immunogenicity level of the generated sequences, we need to first generate 9-mer peptides for each generated sequences
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/FULL_GENERATED_LOW_IMMUNO_SEQUENCES.txt
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/FULL_GENERATED_INTERMEDIATE_IMMUNO_SEQUENCES.txt
python scripts/netmhcpan/generate_peptides_from_sequences.py \
--sequences_filepath=PATH/TO/FULL_GENERATED_HIGH_IMMUNO_SEQUENCES.txt
This run will create 3 .pep files (low, intermediate, and high). Next, we need to run the netMHCpan scoring script:
cd scripts/netmhcpan/
bash netmhcpan_allele_scores.sh PATH/TO/PEP_FILES_DIRECTORY/
This run will create a combined.pep file (in the same directory) which contains the concatenation of low, intermediate, and high peptides (in that order). The combined.pep file will then be passed to the netMHCpan computation. This will results in 12 MHC files that are scored.
Notice that this run is different from the train-valid-test data preparation as this is designed to concatenate generated sequences across different immunogenicity scores mainly to understand the distribution of each immunogenicity score.
Finally, we need to reconcile the peptides into sequences and calculate the score quantiles:
python scripts/netmhcpan/compute_hits_from_peptides.py \
--sequences_filepath=PATH/TO/GENERATED_SEQUENCES.fasta \
--peptides_dir=PATH/TO/PEPTIDES_DIR/ \
--peptide_file_prefix=combined
Similar to the train-valid-test data preparation process, each peptide file that is generated would have a prefix. In this case, it would be combined.
- Fold the proteins using alphafold.sh provided in scripts folder
- Open PyMol, open the proteins and run:
align wuhan_folded, protein_to_evaluate
where wuhan_alphafold denotes the reference protein and protein_to_evaluate denotes the generated protein.
| Model | Train | Val. |
|---|---|---|
| LSTM-small | 1.019 | 1.021 |
| LSTM-base | 1.016 | 1.018 |
| LSTM-large | 1.016 | 1.019 |
| Vaxformer-small | 1.034 | 1.128 |
| Vaxformer-base | 1.014 | 1.043 |
| Vaxformer-large | 1.013 | 1.014 |
| Model | RMSD (\AA) |
|
|---|---|---|
| Random Mutation | 0.32 |
-2.51 |
| Naive Bayes | -0.5 |
|
| VAE | 0.48 |
-5.17 |
| LSTM-base | 0.51 |
-4.95 |
| Vaxformer-large | 0.67 |
-5.45 |
| Model | T-statistics | U-statistics | ||||
|---|---|---|---|---|---|---|
| Medium-Low | High-Medium | High-Low | Medium-Low | High-Medium | High-Low | |
| VAE | 15.22 | 27.68 | 39.17 | 3.99 × 105 | 4.73 × 105 | 5.23 × 105 |
| LSTM-base | 14.23 | 16.83 | 29.75 | 4.45 × 105 | 4.37 × 105 | 5.16 × 105 |
| Vaxformer-large | 26.30 | 38.02 | 56.62 | 4.75 × 105 | 5.21 × 105 | 5.56 × 105 |