The brms package provides an interface to fit Bayesian generalized (non-)linear multivariate multilevel models using Stan, which is a C++ package for performing full Bayesian inference (see http://mc-stan.org/). The formula syntax is very similar to that of the package lme4 to provide a familiar and simple interface for performing regression analyses. A wide range of response distributions are supported, allowing users to fit – among others – linear, robust linear, count data, survival, response times, ordinal, zero-inflated, and even self-defined mixture models all in a multilevel context. Further modeling options include non-linear and smooth terms, auto-correlation structures, censored data, missing value imputation, and quite a few more. In addition, all parameters of the response distribution can be predicted in order to perform distributional regression. Multivariate models (i.e., models with multiple response variables) can be fit, as well. Prior specifications are flexible and explicitly encourage users to apply prior distributions that actually reflect their beliefs. Model fit can easily be assessed and compared with posterior predictive checks, cross-validation, and Bayes factors.
- Introduction to brms (Journal of Statistical Software)
- Advanced multilevel modeling with brms (The R Journal)
- Website (Website of brms with documentation and vignettes)
- Blog posts (List of blog posts about brms)
- Ask a question (Stan Forums on Discourse)
- Open an issue (GitHub issues for bug reports and feature requests)
library(brms)As a simple example, we use poisson regression to model the seizure
counts in epileptic patients to investigate whether the treatment
(represented by variable Trt) can reduce the seizure counts and
whether the effect of the treatment varies with the (standardized)
baseline number of seizures a person had before treatment (variable
zBase). As we have multiple observations per person, a group-level
intercept is incorporated to account for the resulting dependency in the
data.
fit1 <- brm(count ~ zAge + zBase * Trt + (1|patient),
data = epilepsy, family = poisson())The results (i.e., posterior samples) can be investigated using
summary(fit1)
#> Family: poisson
#> Links: mu = log
#> Formula: count ~ zAge + zBase * Trt + (1 | patient)
#> Data: epilepsy (Number of observations: 236)
#> Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
#> total post-warmup samples = 4000
#>
#> Group-Level Effects:
#> ~patient (Number of levels: 59)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 0.58 0.07 0.46 0.73 1.01 757 1812
#>
#> Population-Level Effects:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> Intercept 1.77 0.12 1.54 2.01 1.01 719 946
#> zAge 0.10 0.08 -0.07 0.27 1.00 778 1318
#> zBase 0.71 0.12 0.47 0.95 1.01 593 1323
#> Trt1 -0.27 0.17 -0.61 0.05 1.01 756 1270
#> zBase:Trt1 0.05 0.16 -0.26 0.37 1.01 764 1131
#>
#> Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
#> is a crude measure of effective sample size, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).On the top of the output, some general information on the model is given, such as family, formula, number of iterations and chains. Next, group-level effects are displayed seperately for each grouping factor in terms of standard deviations and (in case of more than one group-level effect per grouping factor; not displayed here) correlations between group-level effects. On the bottom of the output, population-level effects (i.e. regression coefficients) are displayed. If incorporated, autocorrelation effects and family specific parameters (e.g. the residual standard deviation ‘sigma’ in normal models) are also given.
In general, every parameter is summarized using the mean (‘Estimate’)
and the standard deviation (‘Est.Error’) of the posterior distribution
as well as two-sided 95% credible intervals (‘l-95% CI’ and ‘u-95% CI’)
based on quantiles. We see that the coefficient of Trt is negative
with a zero overlapping 95%-CI. This indicates that, on average, the
treatment may reduce seizure counts by some amount but the evidence
based on the data and applied model is not very strong and still
insufficient by standard decision rules. Further, we find little
evidence that the treatment effect varies with the baseline number of
seizures.
The last two values (‘Eff.Sample’ and ‘Rhat’) provide information on how well the algorithm could estimate the posterior distribution of this parameter. If ‘Rhat’ is considerably greater than 1, the algorithm has not yet converged and it is necessary to run more iterations and / or set stronger priors.
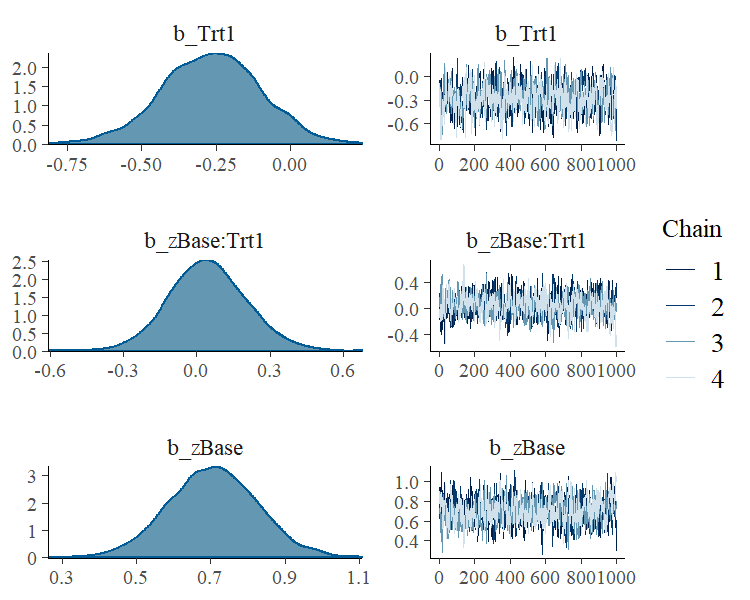
To visually investigate the chains as well as the posterior
distributions, we can use the plot method. If we just want to see
results of the regression coefficients of Trt and zBase, we go for
plot(fit1, pars = c("Trt", "zBase")) A more detailed investigation can be performed by running
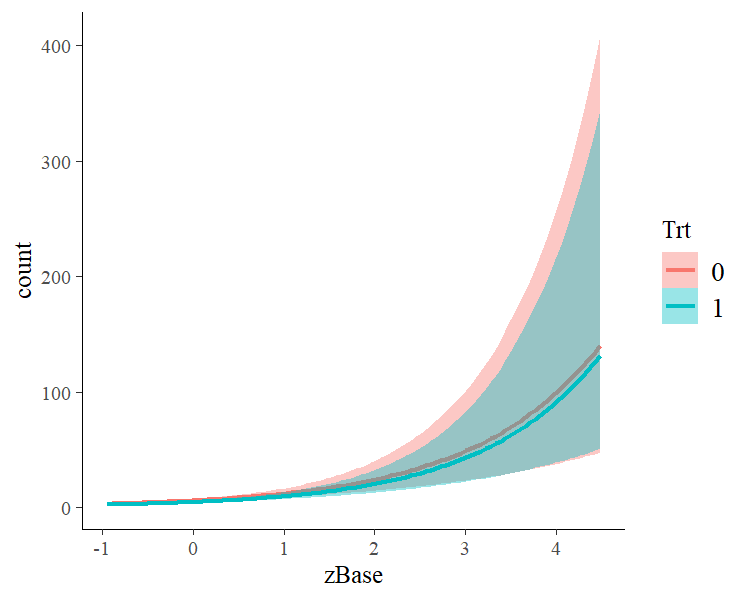
launch_shinystan(fit1). To better understand the relationship of the
predictors with the response, I recommend the conditional_effects
method:
plot(conditional_effects(fit1, effects = "zBase:Trt"))This method uses some prediction functionality behind the scenes, which
can also be called directly. Suppose that we want to predict responses
(i.e. seizure counts) of a person in the treatment group (Trt = 1) and
in the control group (Trt = 0) with average age and average number of
previous seizures. Than we can use
newdata <- data.frame(Trt = c(0, 1), zAge = 0, zBase = 0)
predict(fit1, newdata = newdata, re_formula = NA)
#> Estimate Est.Error Q2.5 Q97.5
#> [1,] 5.92175 2.487403 2 11
#> [2,] 4.57475 2.168084 1 9We need to set re_formula = NA in order not to condition of the
group-level effects. While the predict method returns predictions of
the responses, the fitted method returns predictions of the regression
line.
fitted(fit1, newdata = newdata, re_formula = NA)
#> Estimate Est.Error Q2.5 Q97.5
#> [1,] 5.932893 0.6985409 4.663774 7.447649
#> [2,] 4.545819 0.5293890 3.574081 5.647261Both methods return the same estimate (up to random error), while the
latter has smaller variance, because the uncertainty in the regression
line is smaller than the uncertainty in each response. If we want to
predict values of the original data, we can just leave the newdata
argument empty.
Suppose, we want to investigate whether there is overdispersion in the model, that is residual variation not accounted for by the response distribution. For this purpose, we include a second group-level intercept that captures possible overdispersion.
fit2 <- brm(count ~ zAge + zBase * Trt + (1|patient) + (1|obs),
data = epilepsy, family = poisson())We can then go ahead and compare both models via approximate leave-one-out (LOO) cross-validation.
loo(fit1, fit2)
#> Output of model 'fit1':
#>
#> Computed from 4000 by 236 log-likelihood matrix
#>
#> Estimate SE
#> elpd_loo -672.1 36.5
#> p_loo 94.5 14.2
#> looic 1344.2 73.1
#> ------
#> Monte Carlo SE of elpd_loo is NA.
#>
#> Pareto k diagnostic values:
#> Count Pct. Min. n_eff
#> (-Inf, 0.5] (good) 209 88.6% 368
#> (0.5, 0.7] (ok) 19 8.1% 128
#> (0.7, 1] (bad) 6 2.5% 35
#> (1, Inf) (very bad) 2 0.8% 5
#> See help('pareto-k-diagnostic') for details.
#>
#> Output of model 'fit2':
#>
#> Computed from 4000 by 236 log-likelihood matrix
#>
#> Estimate SE
#> elpd_loo -594.9 13.8
#> p_loo 107.5 6.9
#> looic 1189.8 27.5
#> ------
#> Monte Carlo SE of elpd_loo is NA.
#>
#> Pareto k diagnostic values:
#> Count Pct. Min. n_eff
#> (-Inf, 0.5] (good) 84 35.6% 675
#> (0.5, 0.7] (ok) 100 42.4% 209
#> (0.7, 1] (bad) 45 19.1% 42
#> (1, Inf) (very bad) 7 3.0% 11
#> See help('pareto-k-diagnostic') for details.
#>
#> Model comparisons:
#> elpd_diff se_diff
#> fit2 0.0 0.0
#> fit1 -77.2 27.1The loo output when comparing models is a little verbose. We first see
the individual LOO summaries of the two models and then the comparison
between them. Since higher elpd (i.e., expected log posterior density)
values indicate better fit, we see that the model accounting for
overdispersion (i.e., fit2) fits substantially better. However, we
also see in the individual LOO outputs that there are several
problematic observations for which the approximations may have not have
been very accurate. To deal with this appropriately, we need to fall
back to other methods such as reloo or kfold but this requires the
model to be refit several times which takes too long for the purpose of
a quick example. The post-processing methods we have shown above are
just the tip of the iceberg. For a full list of methods to apply on
fitted model objects, type methods(class = "brmsfit").
Developing and maintaining open source software is an important yet often underappreciated contribution to scientific progress. Thus, whenever you are using open source software (or software in general), please make sure to cite it appropriately so that developers get credit for their work.
When using brms, please cite one or more of the following publications:
- Bürkner P. C. (2017). brms: An R Package for Bayesian Multilevel Models using Stan. Journal of Statistical Software. 80(1), 1-28. doi.org/10.18637/jss.v080.i01
- Bürkner P. C. (2018). Advanced Bayesian Multilevel Modeling with the R Package brms. The R Journal. 10(1), 395-411. doi.org/10.32614/RJ-2018-017
As brms is a high-level interface to Stan, please additionally cite Stan:
- Carpenter B., Gelman A., Hoffman M. D., Lee D., Goodrich B., Betancourt M., Brubaker M., Guo J., Li P., and Riddell A. (2017). Stan: A probabilistic programming language. Journal of Statistical Software. 76(1). 10.18637/jss.v076.i01
Further, brms relies on several other R packages and, of course, on R
itself. To find out how to cite R and its packages, use the citation
function. There are some features of brms which specifically rely on
certain packages. The rstan package together with Rcpp makes
Stan conveniently accessible in R. Visualizations and
posterior-predictive checks are based on bayesplot and ggplot2.
Approximate leave-one-out cross-validation using loo and related
methods is done via the loo package. Marginal likelihood based
methods such as bayes_factor are realized by means of the
bridgesampling package. Splines specified via the s and t2
functions rely on mgcv. If you use some of these features, please
also consider citing the related packages.
To install the latest release version from CRAN use
install.packages("brms")The current developmental version can be downloaded from github via
if (!requireNamespace("remotes")) {
install.packages("remotes")
}
remotes::install_github("paul-buerkner/brms")Because brms is based on Stan, a C++ compiler is required. The program Rtools (available on https://cran.r-project.org/bin/windows/Rtools/) comes with a C++ compiler for Windows. On Mac, you should install Xcode. For further instructions on how to get the compilers running, see the prerequisites section on https://github.com/stan-dev/rstan/wiki/RStan-Getting-Started.
Detailed instructions and case studies are given in the package’s
extensive vignettes. See vignette(package = "brms") for an overview.
For documentation on formula syntax, families, and prior distributions
see help("brm").
Questions can be asked on the Stan forums on Discourse. To propose a new feature or report a bug, please open an issue on GitHub.
If you have already fitted a model, just apply the stancode method on
the fitted model object. If you just want to generate the Stan code
without any model fitting, use the make_stancode function.
When you fit your model for the first time with brms, there is currently
no way to avoid compilation. However, if you have already fitted your
model and want to run it again, for instance with more samples, you can
do this without recompilation by using the update method. For more
details see help("update.brmsfit").
The rstanarm package is similar to brms in that it also allows to fit
regression models using Stan for the backend estimation. Contrary to
brms, rstanarm comes with precompiled code to save the compilation time
(and the need for a C++ compiler) when fitting a model. However, as brms
generates its Stan code on the fly, it offers much more flexibility in
model specification than rstanarm. Also, multilevel models are currently
fitted a bit more efficiently in brms. For detailed comparisons of brms
with other common R packages implementing multilevel models, see
vignette("brms_multilevel") and vignette("brms_overview").