infectio_mesa
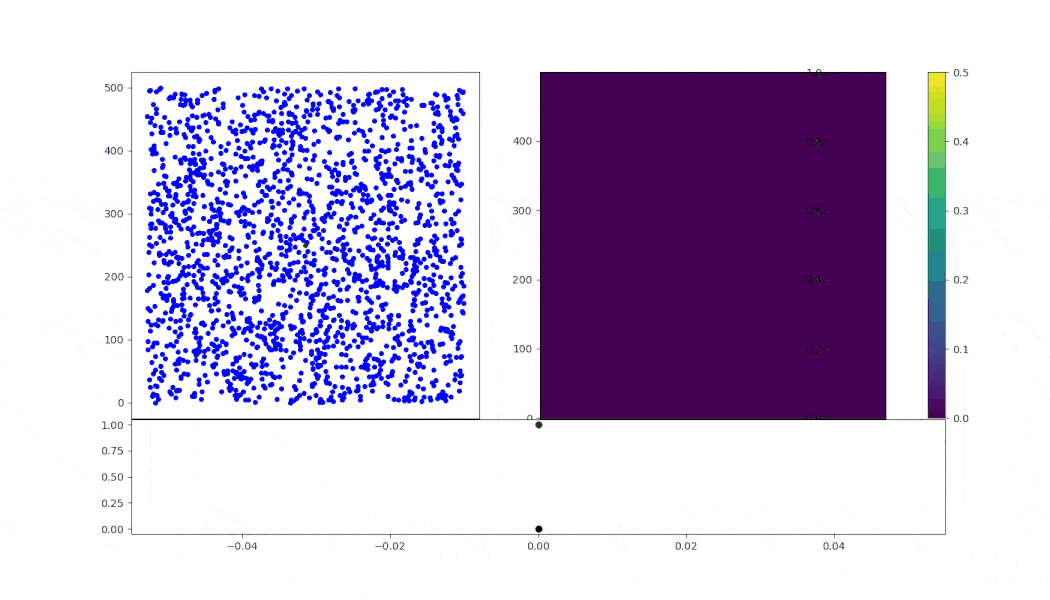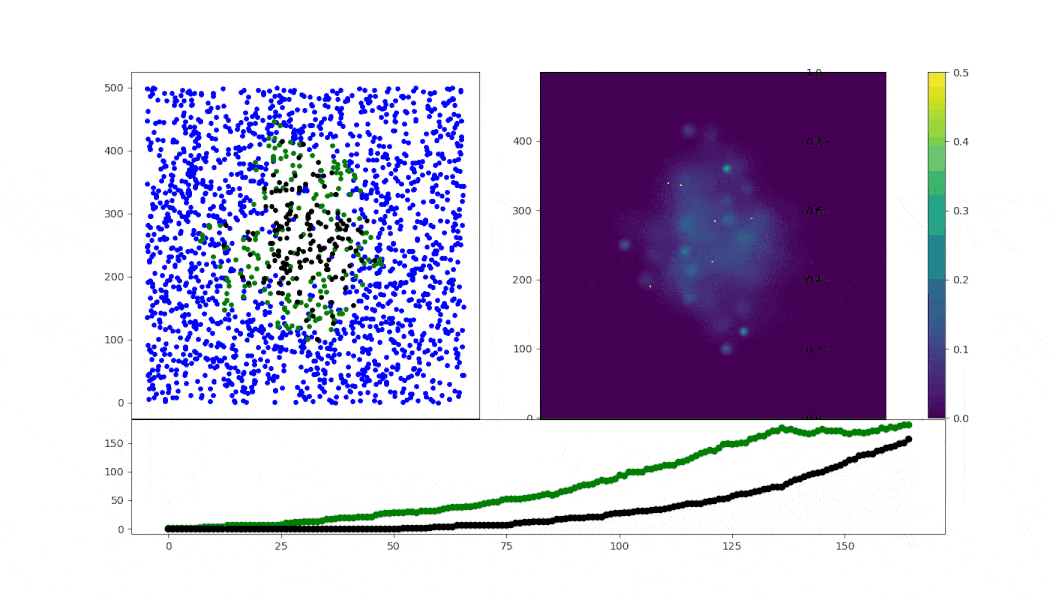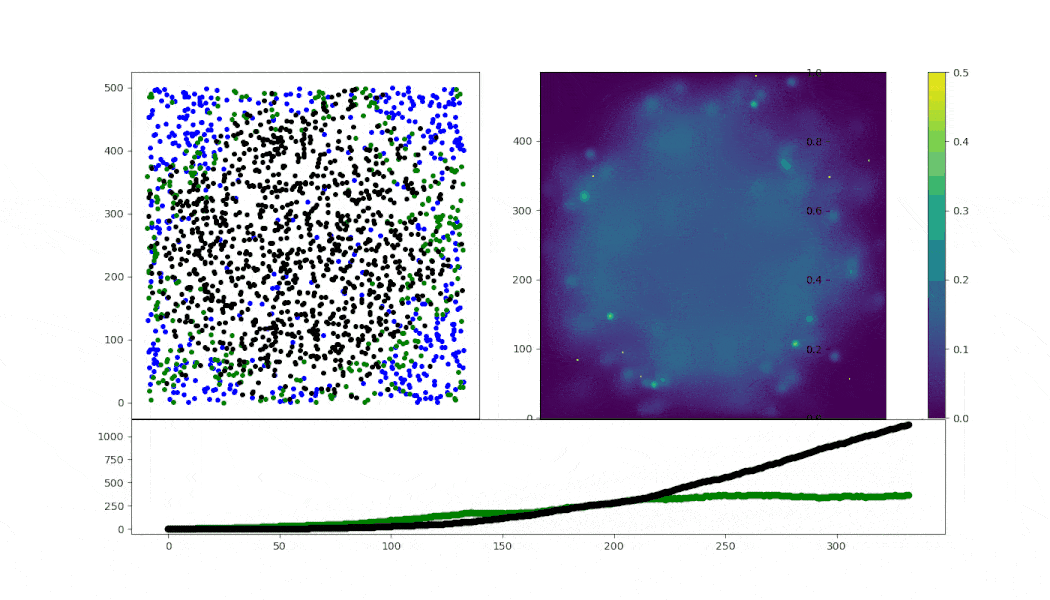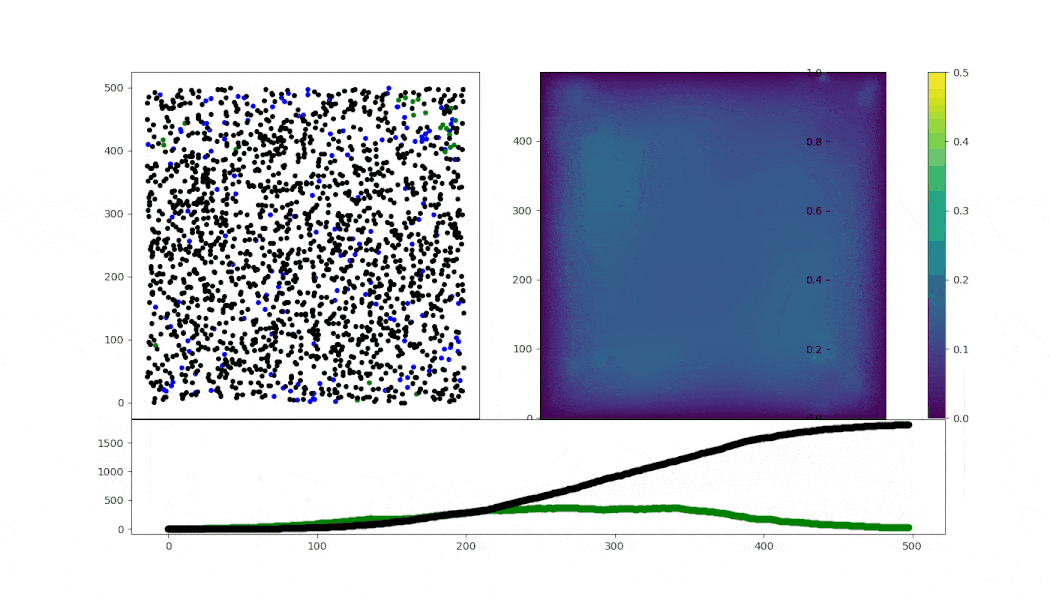
Virus spread simulation using ABM (in mesa) implementing simple Random walk cell motility.
Or:
python run.pypython run_with_matplotlib.py
Running with naive assumptions:
- motility: RW with fixed speed conditioned on state
- cell-cell infection: arbitrary numbers for a sigmoid probability model
- cell death: after 50 steps of infection
- lysis (right now, only fixed interval death, and fixed virions release)
- molecular diffusion: in diffusion.py, clean it and ship it better and evaluate it.
- Put all hyperparameters in a toml/json file
- code to extract parameters and behaviors from dataset
- Remove dependency on Mesa, it is basically not doing any good for this project
- Consider Julia instead of Python if faster needed
- Some ticks showing in plot next to colorbar, remove them
- Saving experiment run
- reproducability