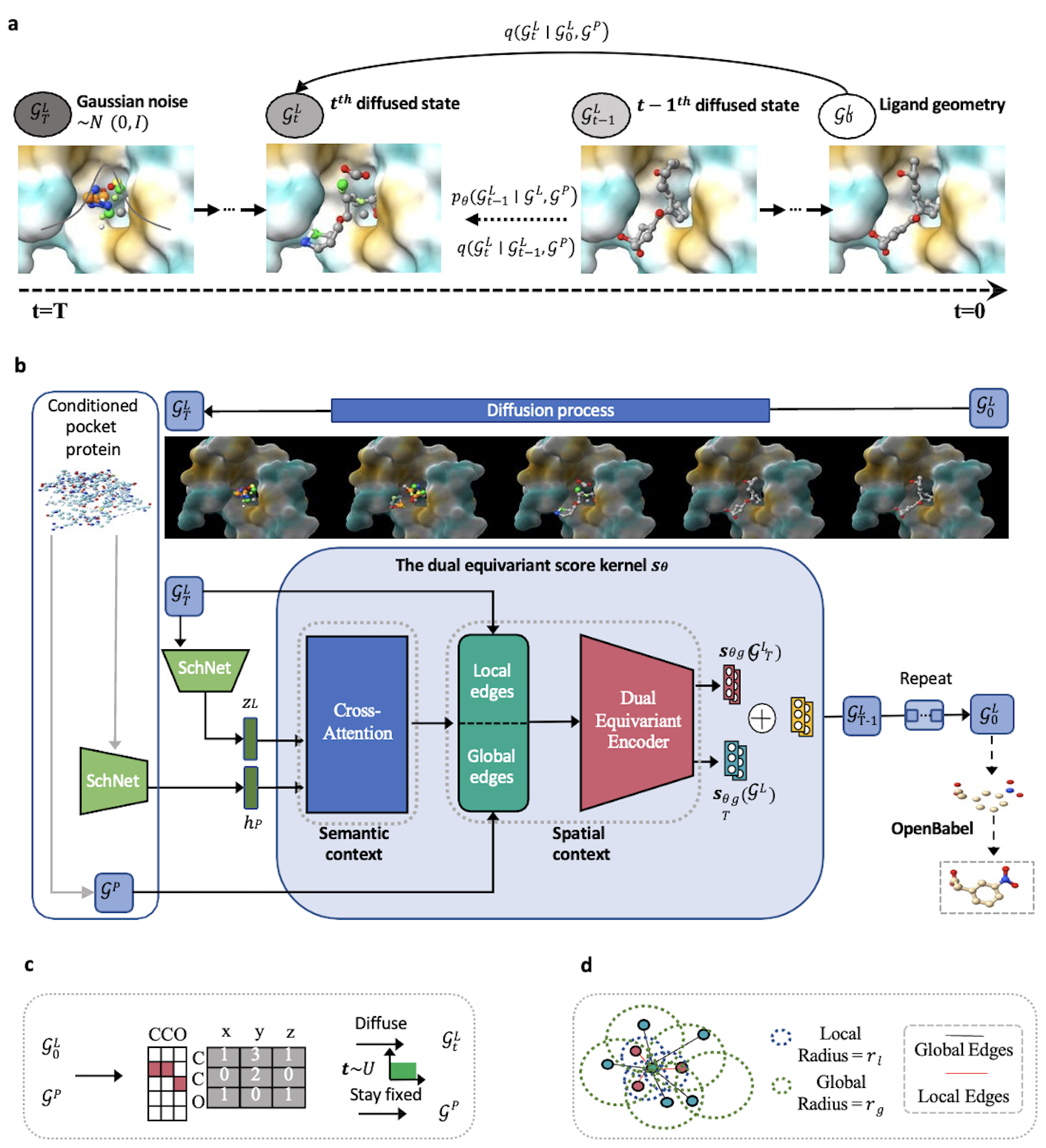
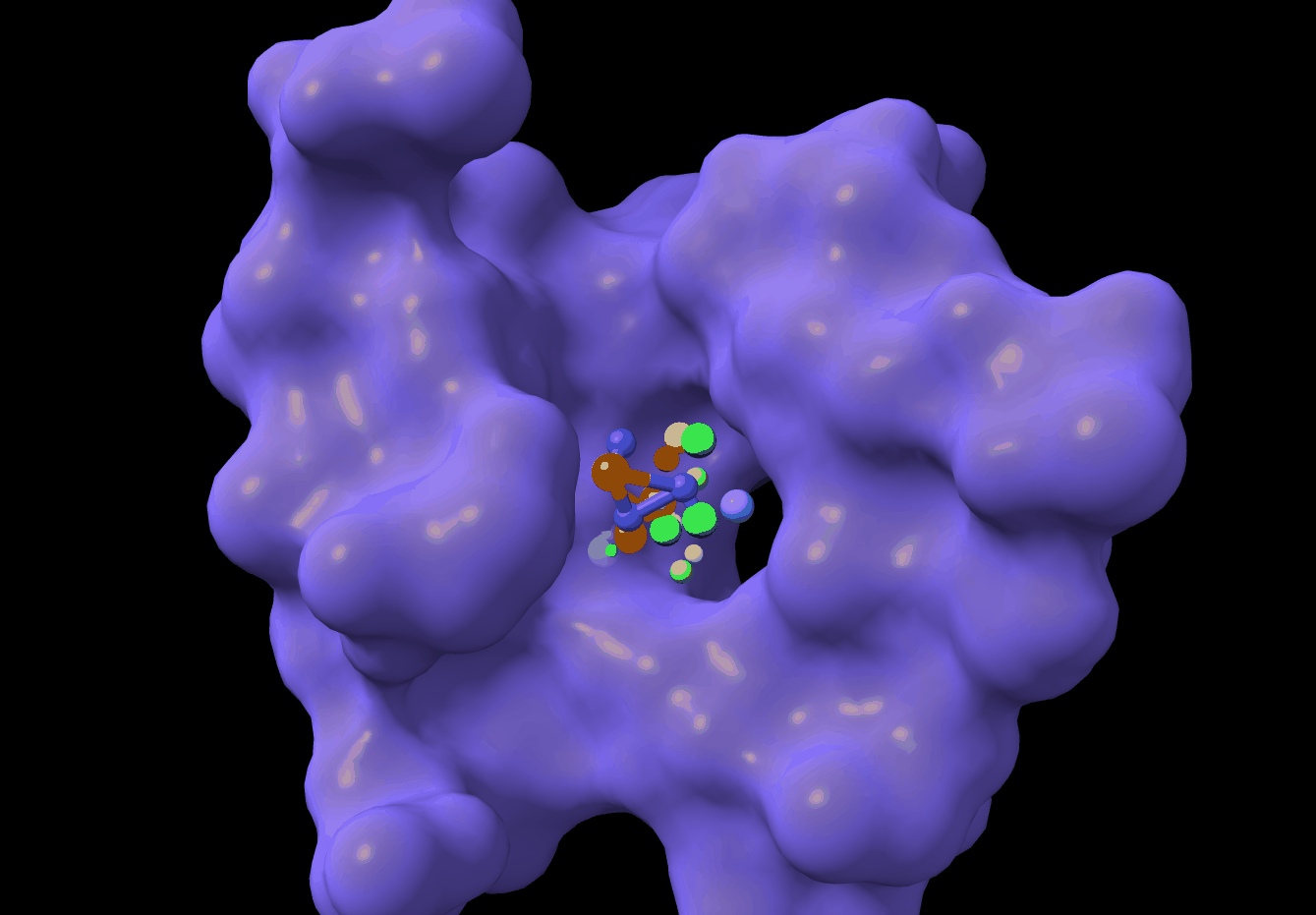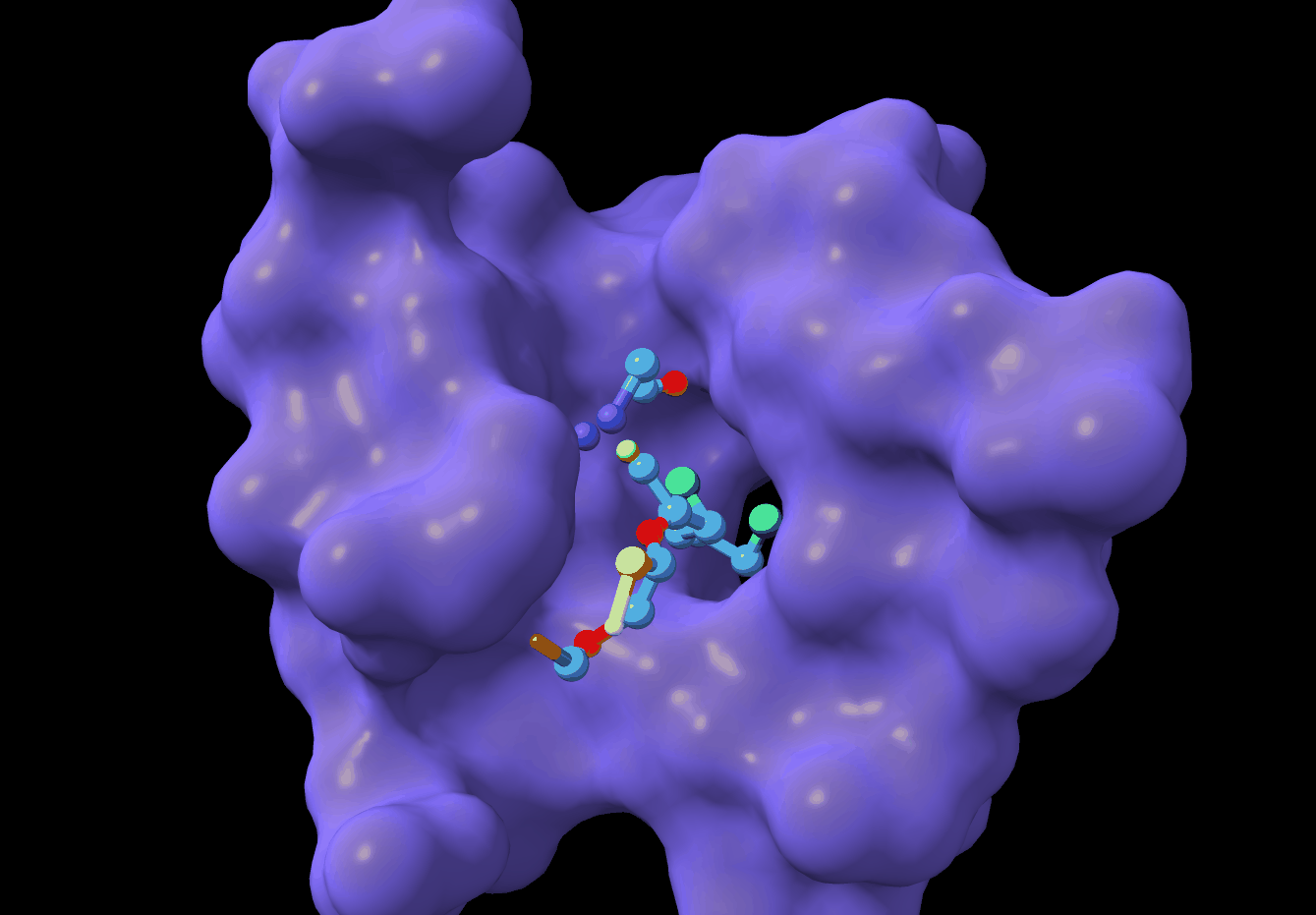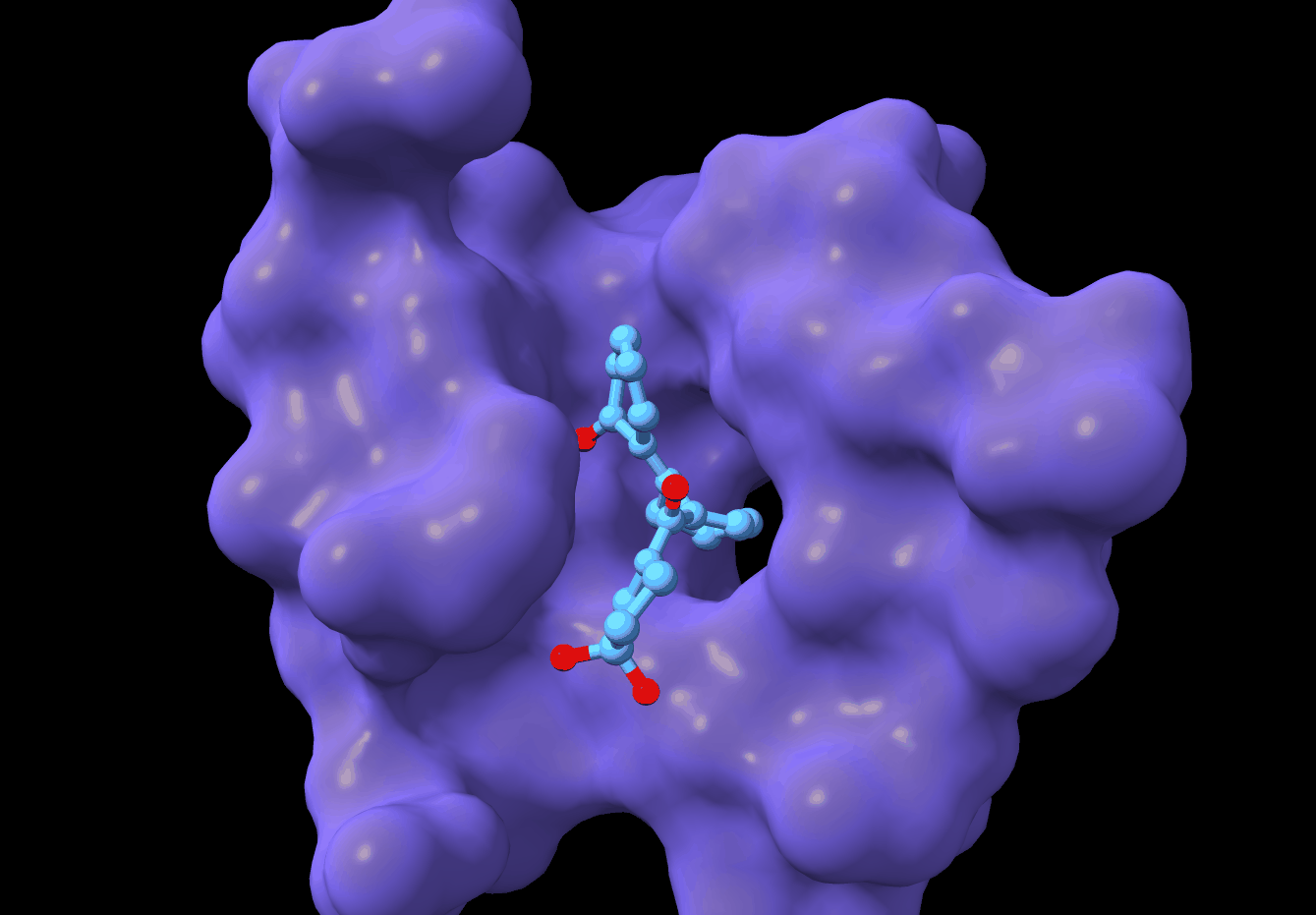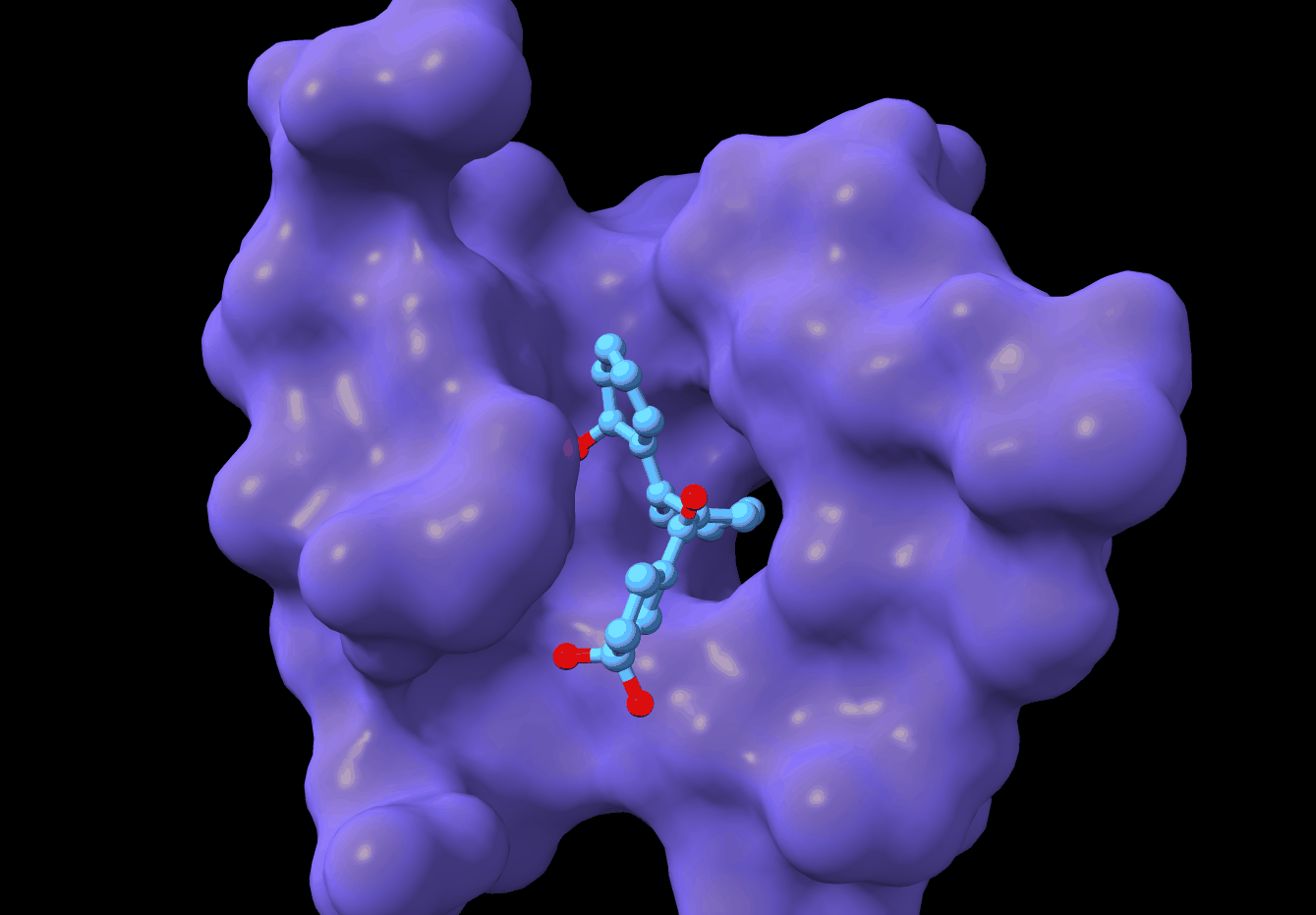
PMDM: A dual diffusion model enables 3D binding bioactive molecule generation and lead optimization given target pockets
Official implementation of PMDM, a dual diffusion model enables 3D binding bioactive molecule generation and lead optimization given target pockets, by Lei Huang.
- Our paper is accepted by Nature Communications !! (https://doi.org/10.1038/s41467-024-46569-1)
Please use our environment file to install the environment.
# Clone the environment
conda env create -f mol.yml
# Activate the environment
conda activate molFor docking, install QuickVina 2:
wget https://github.com/QVina/qvina/raw/master/bin/qvina2.1
chmod +x qvina2.1Preparing the receptor for docking (pdb -> pdbqt) requires a new environment which is based on python 2x, so we need to create a new environment:
# Clone the environment
conda env create -f evaluation/env_adt.yml
# Activate the environment
conda activate adtThe pre-trained models could be downloaded from Zenodo.
Download and extract the dataset is provided in Zenodo
The original CrossDocked dataset can be found at https://bits.csb.pitt.edu/files/crossdock2020/
Download the dataset
wget http://www.bindingmoad.org/files/biou/every_part_a.zip
wget http://www.bindingmoad.org/files/biou/every_part_b.zip
wget http://www.bindingmoad.org/files/csv/every.csv
unzip every_part_a.zip
unzip every_part_b.zipWe provide two training scripts train.py and train_ddp_op.py for single-GPU training and multi-GPU training.
Starting a new training run:
python -u train.py --config <config>.ymlThe example configure file is in configs/crossdock_epoch.yml
Resuming a previous run:
python -u train.py --config <configure file path>The config argument should be the upper path of the configure file.
python -u sample_batch.py --ckpt <checkpoint> --num_samples <number of samples> --sampling_type generalizedpython -u sample_for_pdb.py --ckpt <checkpoint> --pdb_path <pdb path> --num_atom <num atom> --num_samples <number of samples> --sampling_type generalizednum_atom is the number of atoms of generated molecules.
python -u sample_frag.py --ckpt <checkpoint> --pdb_path <pdb path> --mol_file <mole file> --keep_index <seed fragments index> --num_atom <num atom> --num_samples <number of samples> --sampling_type generalizednum_atom is the number of atoms of generated fragments. keep_index is the index of the atoms of the seed fragments.
python -u sample_linker.py --ckpt <checkpoint> --pdb_path <pdb path> --mol_file <mole file> --keep_index <seed fragments index> --num_atom <num atom> --num_samples <number of samples> --sampling_type generalizednum_atom is the number of atoms of generated fragments. mask is the index of the linker that you would like to replace in the original molecule.
Evaluate the batch of generated molecules (You need to turn on the save_results arguments in sample* scripts)
python -u evaluate --path <molecule_path>If you want to evaluate a single molecule, use evaluate_single.py.
First, convert all protein PDB files to PDBQT files using adt envrionment.
conda activate adt
prepare_receptor4.py -r {} -o {}
cd evaluationThen, compute QuickVina scores:
conda deactivate
conda activate mol
python docking_2_single.py --receptor_file <prepapre_receptor4_outdir> --sdf_file <sdf file> --out_dir <qvina_outdir>!!! You have to replace the path of your own mol and adt environment paths with the path in the scripts already.
@article {Huang2023.01.28.526011,
author = {Lei Huang and Tingyang Xu and Yang Yu and Peilin Zhao and Ka-Chun Wong and Hengtong Zhang},
title = {A dual diffusion model enables 3D binding bioactive molecule generation and lead optimization given target pockets},
elocation-id = {2023.01.28.526011},
year = {2023},
doi = {10.1101/2023.01.28.526011},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2023/01/30/2023.01.28.526011},
eprint = {https://www.biorxiv.org/content/early/2023/01/30/2023.01.28.526011.full.pdf},
journal = {bioRxiv}
}