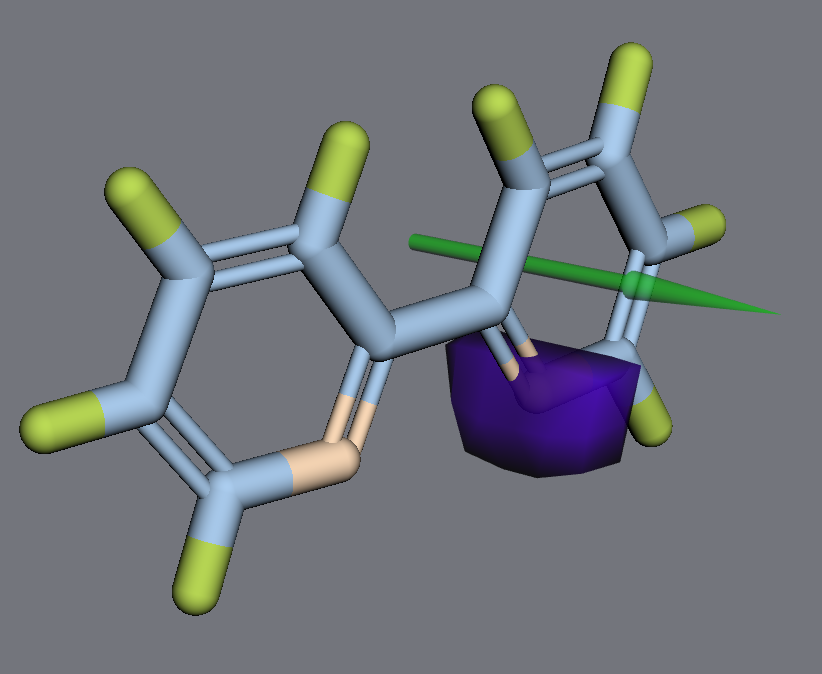
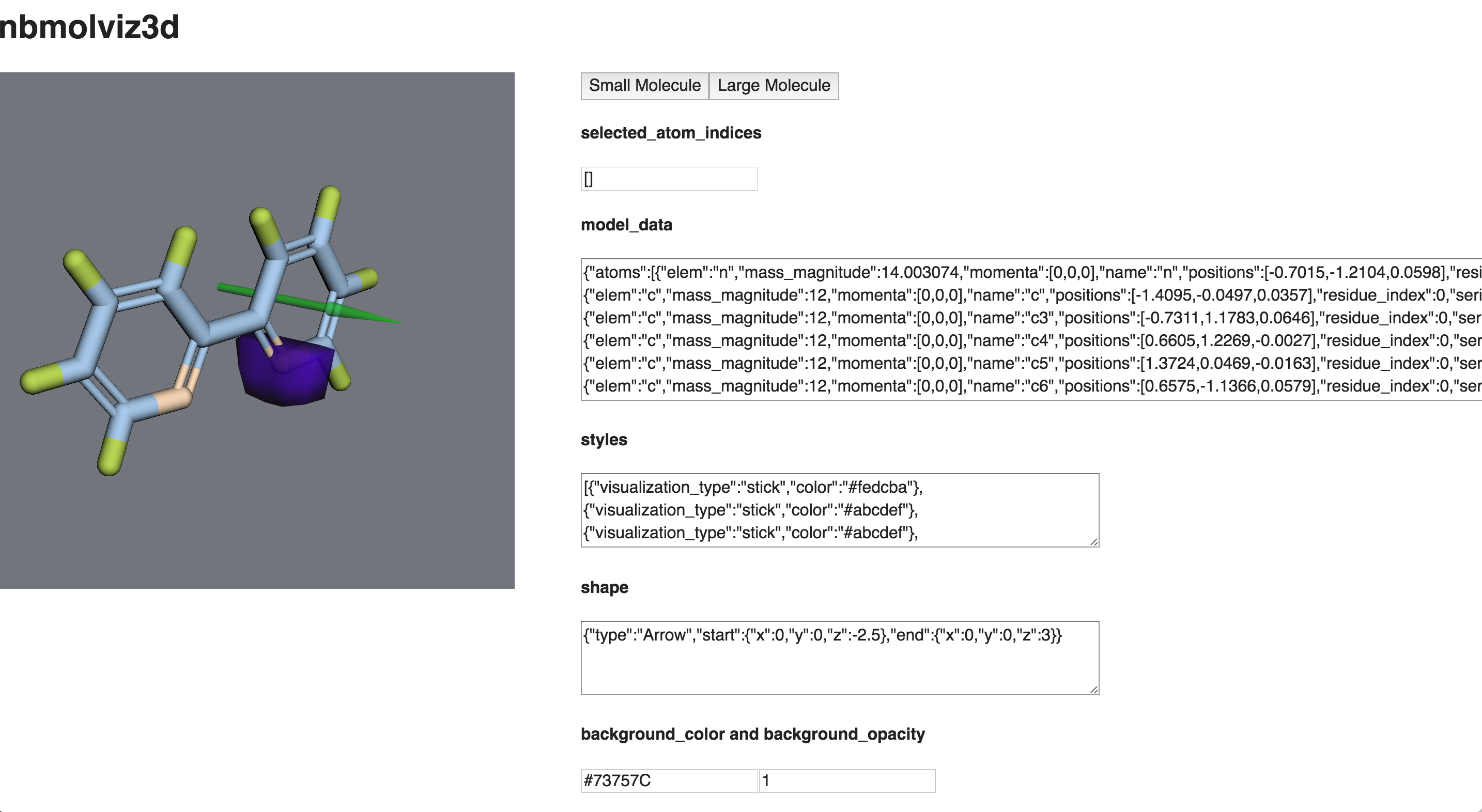
A data-bound React wrapper for 3dmol.js that visualizes any molecule in 3D.
npm install molecule-3d-for-react
<Molecule3d
modelData={{
atoms: [...],
bonds: [...],
}}
/>
See below for the full spec of what data can be passed in, and check out example/js/main.js for a working example.
In order to set up your molecule visualization, just pass in the proper props data to the React component. Here are all of the parameters with explanations:
JSON data representing the actual molecular input. Of the form:
{
atoms: [{
serial,
name,
elem,
mass_magnitude,
residue_index,
esidue_name,
chain,
positions,
momenta,
}, ... ],
bonds: [{
atom1_index,
atom2_index,
bond_order,
}, ... ],
}
An example of full working modelData for a real molecule can be found in example/js/bipyridine_model_data.js.
The background color of the visualization.
The opacity of the background.
Indicates whether or not to show text labels on all atoms.
An array indicating how to style individual atoms. Atoms are indicated by index, so the first style in this array corresponds to the first atom in model_data.atoms. Of the form:
[
{
visualization_type: 'stick'|'sphere'|'cartoon',
color: '#abcdef',
}, ...
]
An example of a styles array for the bipyridine molecule can be found in example/js/bipyridine_styles.js.
An array of atom indices indicating which atoms should be visually selected.
A string indicating whether clicks select atoms ('Atom'), residues ('Residue'), or chains ('Chain').
Indicates any shapes to display in the visualization using 3Dmol.js's addShape method. For example:
[{
type: 'Sphere',
x: 0,
y: 0,
z: 0,
}]
Labels to draw using 3Dmol.js's addLabel method. The text for the field is expected as label.text. For example:
[{'backgroundColor': '0x000000',
'backgroundOpacity': 1.0,
'borderColor': 'black',
'fontColor': '0xffffff',
'fontSize': 14,
'position':
{'x': -1.84,
'y': 8.30,
'z': 33.87},
'text': 'PRO1'}]Indicates an orbital to display using 3Dmol.js's addIsosurface method. Of the type:
{
cube_file,
iso_val,
opacity,
}
A callback for when the modelData has changed and it has been re-rendered in the viewer.
An example is included which provides data-bound inputs that you can play with to see how they affect the visualization. To run it, use the command:
npm run example
Take a look at our sister project, molecule-2d-for-react, for a React component with a similar interface that renders a 2d visualization.
Running the example above will also set up a typical development flow, where any changes to the code will be immediately reflected in the browser.
If you're using this in another project and want to make changes to this repository locally and see them reflected in your other project, first you'll need to do some setup. You can point your other project to use the local copy of molecule-3d-for-react like this:
cd ~/path/to/molecule-3d-for-react
npm link
cd ~/path/to/other-project
npm link molecule-3d-for-react
See this great blog post for more info on npm link.
Once you've linked your other project, you'll need to build molecule-3d-for-react (and likely your other project, too) every time you want your changes to reflect in your other project. You can do this manually with npm run build. If you want to rebuild molecule-3d-for-react automatically every time a change is made, run npm run watch.
Unit tests can be run with:
npm test
End-to-end tests can be run with:
npm run setup-selenium
npm run e2e
Travis automatically publishes any new tagged commit to NPM. The best way to take advantage of this is to first create a new tagged commit using npm version:
npm version patch -m "Upgrade to %s for reasons"
Then push that commit to a new release branch, push the tag with git push origin --tags and open a pull request on Github. When you see that Travis has succeeded in deploying, merge it to master.
Copyright 2016 Autodesk Inc.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
This project is developed and maintained by the Molecular Design Toolkit project. Please see that project's CONTRIBUTING document for details.