note The script findRBBHs.pl uses the -max-target-seqs flag in blast, which is technically incorrect because it does not always necessarily return the best hit. I believe this shouldn't have a large impact here because the search databases are so small.
This repository holds the code used to analyze data from an exon capture experiment.
The general strategy is as follows:
- Quality control data (remove reads not passing CASAVA filter, remove adapter contamination, window-trim for sequence quality, merge overlapping reads)
- Combine data for all of the libraries for a single species into a single set of fastq files
- Perform targeted assemblies of our exon data using the ARC assembly pipeline from iBest
- Map reads from individual libraries back to the assemblies made with ARC to assess how different laboratory conditions affected capture efficiency
The pipeline is run from a driver script called runAnalyses.pl. You must provide it with a configuration file as follows: perl runAnalyses.pl -c configurationFile.txt. The configuration file contains information regarding the names of the sample libraries and where they are located. Its form should be as follows (it includes one header row and should contain no whitespace):
AdapterFolder=./adapters
ReadsFolder=./concatenatedReads
QCoutputFolder=./qc
ARCworkingFolder=/home/evan/ramdisk
threads=30
targets=targets.fasta
cycles=6
timeout=180
Sample=SampleID1
Sample=SampleID2
Sample=SampleID3
...
Sample=SampleIDlast
The manderCap repository should be in the same directory as the directories containing the read data (in this case those directories are called Project_Shaffer and Undetermined_indices). We'll call this top-level directory that contains the manderCap repo and the Project_Shaffer and Undetermined_indeces folders "topDirectory" from here on out.
Move all the reads from their Sample folders into a single directory:
cd topDirectory
mkdir allReads
mv Project_Shaffer/Sample_0*/*.fastq.gz allReads/
We don't need the empty sample folders, Sample Sheets, or Basecall_Stats. We also won't use the reads that weren't demultiplexed. So nuke those:
rm -r Project_Shaffer
rm -r Undetermined_indices
Now we want to make a single R1 and single R2 file for each sample. We'll put those into a new directory:
mkdir concatenatedReads
cd allReads
bash ../manderCap/concatenateSampleReads.sh > ../logs/concatenateSampleReads.log 2>&1
We also want the read files to be unzipped:
cd ../concatenatedReads; gunzip *.gz > ../logs/gunzipSampleReads.log 2>&1
Finally, we want to produce the sample-specific adapter FASTAs that we'll use to trim adapter contamination:
cd topDirectory
mkdir adapters
perl manderCap/generate_adapter_fastas.pl --in manderCap/HSEM020_adaptersKey.txt --out adapters --adapters itru > logs/generate_adapter_fastas.log 2>&1
The following command runs all the read QC and ARC assemblies (see that script for details):
perl manderCap/runAnalyses.pl -c manderCap/manderCap.config > logs/runAnalyses.stdout 2>logs/runAnalyses.stderr
After ARC has run, we'll process the ARC assembled contigs to arrive at our final reference that we'll use to map all of our individual libraries to. We first want to designate a single assembled contig that is representative of each target. To do this, we'll find the Reciprocal Best Blast Hits (RBBHs) for each target. This is performed by manderCap/findRBBHs.pl. Briefly, it blasts the contigs in the assembly against the sequences in the target fasta file. Then it blasts the targets in the fasta file against the assembly. Then it goes through these blast reports and finds all instances where the targets had at least one hit to the assembly. For all of the best hits, it checks to make sure that that target was also found as the best hit when we blasted the assembly against the targets (hence the reciprocal part).
cd ARC/finished_allCTS/
cp contigs.fasta contigs.CTSonly6iter.fasta
../../manderCap/findRBBHs.pl --assembly contigs.CTSonly6iter.fasta --targets ../../targets.fasta --out RBBHs.CTSonly6iter.fasta > ../../logs/findRBBHs.log 2>&1
That process finds a total of 8386 reciprocal best blast hits (over 96%). If we were
to map reads to this assembly and visualize coverage, we'd see a fair amount of targets
that have spikes in read coverage at the ends of the targets. This can be the result
of several things. It's possible that these are true repetitive regions that exist
at the edges of our target (at the periphery of the exons, for instance). They may
also be due to chimerism in our assemblies, whereby repetitive, non-contiguous genomic
regions are grafted onto the edges targets due to challenges in de novo assembly.
Here is one example of what what I'm talking about (the black bar represents the portion
of the target that blasted to that assembled contig):
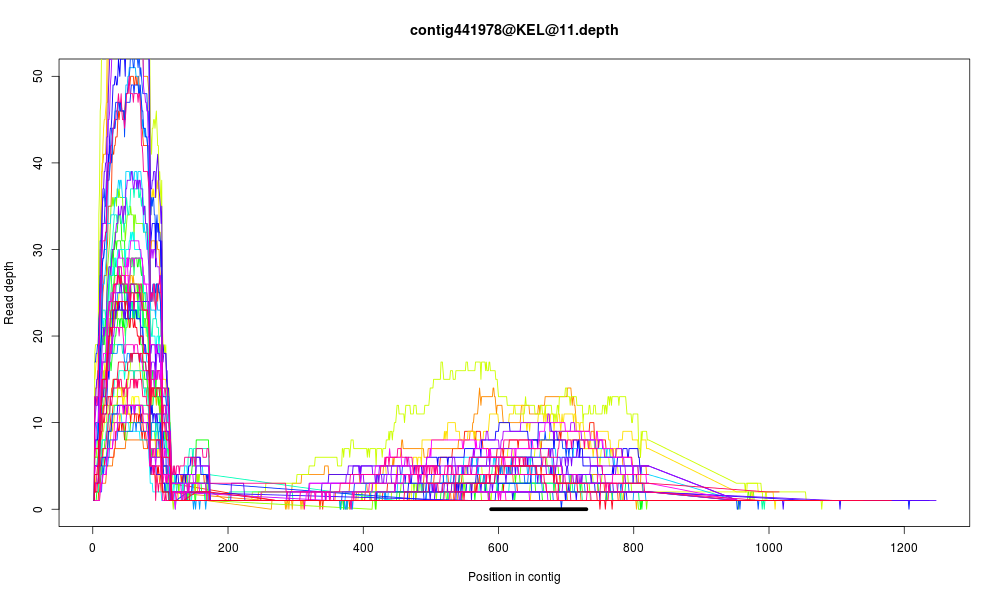
To minimize this effect, we'll try to mask some of these repetitive regions prior to mapping. Since a given repetitive region is typically present in more than one representative target contig, we'll do this by masking regions that blast to multiple targets via self-blast:
makeblastdb -in RBBHs.CTSonly6iter.fasta -dbtype nucl > ../../logs/makeblastdb.log 2>&1
blastn -db RBBHs.CTSonly6iter.fasta -query RBBHs.CTSonly6iter.fasta -out RBBHs.CTSonly6iter.selfBlast -evalue 1e-20 -outfmt 6
perl ../../manderCap/maskChimeras.pl --in RBBHs.CTSonly6iter.selfBlast --sequences RBBHs.CTSonly6iter.fasta --out RBBHs.CTSonly6iter.chimeraMasked.fasta > ../../logs/maskChimeras.log 2>&1
get_fasta_lengths.py --input RBBHs.CTSonly6iter.chimeraMasked.fasta
Reads: 8,386
Bp: 11,813,530
Avg. len: 1,408.72048653
STDERR len: 3.4372379005
Min. len: 210
Max. len: 6,284
Median len: 1,398.0
Contigs > 1kb: 7,816
We'll use this as our assembly for downstream analyses:
cd ../..;mkdir assembly;cp ARC/finished_allCTS/RBBHs.CTSonly6iter.chimeraMasked.fasta assembly/;
bwa index RBBHs.CTSonly6iter.chimeraMasked.fasta > ../logs/indexAssembly.log 2>&1
Now we'll map reads to the assembly we prepared in the previous step. The general strategy is to map single reads, then paired end reads. Then we merge the bam files (samtools), clean them (picard), sort them (picard), and mark duplicates (picard). The MarkDuped files are then analyzed using flagstat (samtools), and depth-calculated (samtools depth). Those depth files will be used to evaluate the individual loci. All of these steps are implemented in manderCap/mapReads.pl.
cd topDir; mkdir mapping; mkdir mapping/depths
perl manderCap/mapReads.pl --config manderCap/manderCap.config --reference /mnt/Data1/HSEM020/assembly/RBBHs.CTSonly6iter.chimeraMasked.fasta > logs/readMapping.log 2>&1
We want to be clear that these bam files and depth files are the result of mapping to the CTS-only 6 ARC iteration assembly. So we'll rename the mapping and depths folders:
mv mapping mappingCTS6iter
cd mappingCTS6iter
mv depths depthsCTS6iter
Generate the locus-specific read depths:
cd depthsCTS6iter
perl ../../manderCap/createLocusDepthFiles.pl --out locusDepthFilesCTSonly6iter > ../../logs/createLocusDepthFiles.CTSonly6iter.log 2>&1
Generate the locus-specific plots of sequencing depth across the loci:
cd locusDepthFilesCTSonly6iter;
ls -al | grep "\.depth" | awk '{print $(NF)}' > dpthFilesCTSonly6iter.txt
mkdir plots
library("data.table")
depthFiles <- fread("depthFiles.txt", sep="\n", header=FALSE)
colors <- c("green", "blue")
# Now depthFiles$V1 holds all of the individual loci files--we'll read them one-by-one and create the plots
for (i in 1:length(depthFiles$V1)) {
dataz <- fread(depthFiles$V1[i])
samples <- unique(dataz$V1)
imageName <- paste("plots/", depthFiles$V1[i], ".png", sep="")
paste("Processing ", depthFiles$V1[i], ". locus: ", depthFiles$V1[i], "\n")
png(filename = imageName, width=1000, height=600, units="px", pointsize=12)
# Make the initial plot
plot(dataz$V3[dataz$V1 == samples[1]], dataz$V4[dataz$V1 == samples[1]], type="l", xlim=c(0,max(dataz$V3)), ylim=c(0,500), col=colors[1], lwd=1.2, main=depthFiles$V1[i], xlab="Position in contig", ylab="Read depth")
# Add all the lines for the rest of the samples
for (i in 2:length(samples)) {
lines(dataz$V3[dataz$V1 == samples[i]], dataz$V4[dataz$V1 == samples[i]], type="l", col=colors[i], lwd=1.5)
}
dev.off()
rm(dataz)
gc()
}
Here we process the depth files generated by samtools depth on the MarkDuped files.
cd ../../../ARC/finished_allCTS/
makeblastdb -in RBBHs.CTSonly6iter.chimeraMasked.fasta -dbtype nucl
blastn -db RBBHs.CTSonly6iter.chimeraMasked.fasta -query ../../targets.fasta -outfmt 6 -evalue 1e-20 -out targets_bl2_chimeraMaskRBBHs.1e-20.blast
cd ../../mappingCTS6iter/depthsCTS6iter
perl ../../manderCap/countSuccessfulTargets.pl --blast ../../ARC/finished_allCTS/targets_bl2_chimeraMaskRBBHs.1e-20.blast --assembly null --depthfiledir ./ --out sampleMetrics > ../../logs/countSuccessfulTargets.log 2>&1
perl ../../manderCap/targetSpecificMetrics.pl --blast ../../ARC/finished_allCTS/targets_bl2_chimeraMaskRBBHs.1e-20.blast --assembly null --depthfiledir ./ --out targetMetrics > ../../logs/targetSpecificMetrics.log 2>&1
cd ../../
These two scripts should output four different files: sampleMetrics.HSPcounts.txt, sampleMetrics.aveDepths.txt, targetMetrics.aveDepths.txt, and targetMetrics.wholeHSPcounts.txt.
The sampleMetrics files contain one row for every sample, with the aveDepths file showing
the average depth across all targets for the whole HSP region and the highest 100bp window
within the HSP region. The targetMetrics files contain one row for every target that had at
least one qualifying read across the HSP region (ang also a header line). The aveDepths file
contains two numbers for each target--the average depth across all samples, and the average
depth of the highest-depth 100bp across all samples. The targetMetrics.wholeHSPcounts.txt file
contains counts of the numbers of samples that had average depth (corrected by sequencing errort)
across the HSP of less than 5, greater than 5, greater than 10, and greater than 20, as well as
the same quantities of the highest-depth 100bp. There are 8,208 such targets represented in the
targetMetrics files.
We only want to include targets that contain at least one qualifying polymorphism across the baited region. To do this, we first need to call SNPs in the targets, and find those targets that have a SNP that is heterozygous in CTS and heterozygous in the F1. Such SNPs are potentially diagnostic between CTS and BTS. First we need to call SNPs using all of the CTS and F1 reads:
mkdir callSNPs
cd callSNPs
cp ../mappingCTS6iter/depthsCTS6iter/targetMetrics.* ./
bash ../manderCap/makeCTSandF1_fastqs.sh > ../logs/makeCTSandF1_fastqs.log # Create the master F1 and CTS fastq files to map
perl ../manderCap/callSNPs.pl > ../logs/callCTSandF1SNPs.log 2>&1
After this we end up with a file containing SNPs: CTSandF1-Q30SNPs.vcf. We'll next pull out all the
SNPs that passed our filters using grep PASS CTSandF1-Q30SNPs.vcf > CTSandF1-Q30SNPs_passOnly_noHeader.vcf.
We then want to sort through these and find only those SNPs where the CTS is homozygous and the F1 is
heterozygous. These SNPs will show the pattern 0/0 0/1 or 1/1 0/1. We can use grep to pull those out as
well: grep -P "(0\/0\:\d.*\t0\/1\:)|(1\/1\:\d.*\t0\/1\:)" CTSandF1-Q30SNPs_passOnly_noHeader.vcf > qualifyingSNPs.vcf
Now that we have a list of qualifying SNPs, we want to look at each target individually, and count how many SNPs occur in the target (baited) region, as defined by the bait set used to enrich these targets:
blastn -db ../ARC/finished_allCTS/RBBHs.CTSonly6iter.chimeraMasked.fasta -query ../targetsNoBd.fasta -outfmt 6 -out targets_bl2_RBBHsChimeraMasked.blast
perl ../manderCap/countSNPsOverTarget.pl --vcf qualifyingSNPs.vcf --blast targets_bl2_RBBHsChimeraMasked.blast > snpPercentages.txt
We only included targets in the average depth calculations that had blast HSPs from the original targets to the assembled contigs that were greater than 100bp long. This left us with 8,208 total targets. When we called SNPs across targets and counted how many SNPs were present across HSPs, we did not give ourselves a 100bp minimum. We found a total of 6,961 targets with at least one SNP that was homozygous in CTS and heterozygous in the F1 (potentially segregating sites between CTS and BTS).
However, 58 of these targets had highest-scoring HSPs that were less than 100bp long,
including one OPA target: OPA2a|E6E9|OPA. So this means that we have a list of 6,901
targets with HSPs at least 100bp long that contain at least one potential ancestry-informative
SNP.
We want to take the intersection of targets that had at least 5bp of depth for the highest 100bp window found within an HSP and targets that had at least 1 potentially segregating CTS/BTS site.
We can do that in R by merging a few results files, but first we need to add a header
row to the SNP percentages file: echo -e "Target\tSNPpercentage" | cat - snpPercentages.txt > snpPercentages_withHeader.txt
targetStatsAve <- read.csv("targetMetrics.aveDepths.txt", sep="\t")
SNPpercentagesAcrossBaits <- read.table("snpPercentages_withHeader.txt", quote="\"", header=TRUE, sep="\t")
targetAveWithSNPcount <- merge(targetStatsAve, SNPpercentagesAcrossBaits, "Target")
goodTargets <- targetAveWithSNPcount[targetAveWithSNPcount$max100AverageTargetDepth >= 5,]
length(goodTargets$SNPpercentage)
# [1] 5260
write.table(goodTargets, file="targets_Max100AveDepthOver5_withSNPs.tsv", sep="\t", row.names=FALSE)Those commands give us a list of 5,260 targets. Now let's take those 5,260 targets and extend their edges out in the assembled contigs if HSP was less than 250bp long. First we need to translate the target names from "allCTS_:contig00003|E19A4|OPA:_Contig001_contig00003|E19A4|OPA" to "contig00003|E19A4|OPA". Run this to do that:
#!/usr/bin/perl
use strict;
use warnings;
open(my $FH, "<", "targets_Max100AveDepthOver5_withSNPs.tsv") or die "Couldn't open targets_Max100AveDepthOver5_withSNPs.tsv for reading: $!\n";
open(my $outFH, ">", "targetsRenamed_Max100AveDepthOver5_withSNPs.tsv") or die "Couldn't open targetsRenamed_Max100AveDepthOver5_withSNPs.tsv for writing: $!\n";
while(my $line = <$FH>) {
if ($line =~ /.*\_\:\_(.*)\_\:\_.*?\t(.*)/) {
print $outFH $1 . "\t" . $2 . "\n";
} else {
print $outFH $line;
}
}cp ../ARC/finished_allCTS/RBBHs.CTSonly6iter.chimeraMasked.fasta ./
perl ../manderCap/extendFinalTargets.pl --targets targetsRenamed_Max100AveDepthOver5_withSNPs.tsv --blast targets_bl2_RBBHsChimeraMasked.blast --assembly RBBHs.CTSonly6iter.chimeraMasked.fasta --minlength 300 --out finalTargetSetDepth5_atleast1SNP_min300bp.fasta
Three targets were dropped because the assembled contigs were less than 300bp long.
Now make a blast database out of that final target set and run a self blast:
makeblastdb -in finalTargetSetDepth5_atleast1SNP_min300bp.fasta -dbtype nucl
blastn -db finalTargetSetDepth5_atleast1SNP_min300bp.fasta -query finalTargetSetDepth5_atleast1SNP_min300bp.fasta -outfmt 6 -out finalTargetSetDepth5_atleast1SNP_min300bp.bl2self.blast
Since we end up with more lines in the self blast output file, we want to find which targets had multiple hits and/or multiple HSPs: ``` perl ../manderCap/flagMultiHits.pl --in finalTargetSetDepth5_atleast1SNP_min300bp.bl2self.blast Multiple hsps found for allCTS_:_contig315752|FAM198A|11_:_Contig001_contig315752|FAM198A|11 Multiple hsps found for allCTS_:_contig315214|SNX33|6_:_Contig002_contig315214|SNX33|6 Multiple hsps found for allCTS_:_contig359783|DERL2|6_:_Contig001_contig359783|DERL2|6 Multiple hsps found for allCTS_:_contig211570|TLCD1|11_:_Contig001_contig211570|TLCD1|11 Multiple hsps found for allCTS_:_contig124104|BET1|8_:_Contig001_contig124104|BET1|8 Multiple hsps found for allCTS_:_contig217149|RYBP|12_:_Contig001_contig217149|RYBP|12 Multiple hsps found for allCTS_:_contig334836|MOCS2|6_:_Contig003_contig334836|MOCS2|6 Multiple hsps found for allCTS_:_contig319555|NARS2|6_:_Contig001_contig319555|NARS2|6 ```
These targets all have multiple HSPs within themselves. This indicates that either the target itself contains a repetitive region or that the assembly somehow included some repetitive sequence. Because none of them are OPAs, and because there are only eight of them, we'll just remove them entirely from the target set:
perl ../manderCap/removeMultiHSPtargets.pl --in finalTargetSetDepth5_atleast1SNP_min300bp.fasta --out finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta
get_fasta_lengths.py --input finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta
Reads: 5,249
Bp: 1,694,651
Avg. len: 322.852162317
STDERR len: 0.498945889134
Min. len: 300
Max. len: 455
Median len: 300.0
Contigs > 1kb: 0
Now we'll check it to make sure there are no multi-hit or multi-HSP blast records.
makeblastdb -in finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta -dbtype nucl
blastn -db finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta -query finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta -out finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.bl2self.blast -outfmt 6
wc -l finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.bl2self.blast
5249 finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.bl2self.blast
Looks good!
The last thing we need to do is deal with the chimera-masked sequence in our data. Recall that prior to read mapping we hard masked sequence regions that had positive blast HSPs to other assembled targets in the assembly (converted those cross-matching target regions to N's). We obviously don't want to design bait sequences with N's in them. We also don't want to do as much as we can to avoid capturing repetitive DNA that flanks target regions, so we may want to avoid those targets entirely. Let's first see how many targets made it into our target set that have N's in them:
grep -cP "^[ATCGN].*N" finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta
16
There are only 16 total lines in our target seqs that were extended into "N" regions (this corresponds to 13 total targets, because three of them had Ns on multiple lines of the sequence file). Of these are OPA targets, the easiest thing to do would be to just remove them all.
Additionally, if these targets received N's in them, it could potentially mean there are repetitive regions very close to the original baited regions. These are difficult to handle from a SNP calling perspective, so we might benefit from not having them. To remove all targets that have any N's in their sequence:
#!/usr/bin/perl
use strict;
use warnings;
use Getopt::Long;
use Bio::SeqIO;
my $help = 0;
my $inFile;
my $outFile;
GetOptions ("in=s" => \$inFile,
"out=s" => \$outFile,
"help|man" => \$help) || die "Couldn't get options with GetOpt::Long: $!\n";
if (!$inFile or !$outFile or $help) {
die "Must supply --in and --out.\n";
}
my $seqIn = Bio::SeqIO->new(-file => $inFile,
-format => 'fasta');
my $seqOut = Bio::SeqIO->new(-file => ">$outFile",
-format => 'fasta');
while (my $seq = $seqIn->next_seq()) {
my $sequence = $seq->seq();
if ($sequence =~ /[nN]/i) {
next;
} else {
$seqOut->write_seq($seq);
}
}After running the above script as perl removeNseqs.pl --in finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP.fasta --out finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta, we can check to see what remains:
get_fasta_lengths.py --input finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta
Reads: 5,236
Bp: 1,690,740
Avg. len: 322.906799083
STDERR len: 0.499950509799
Min. len: 300
Max. len: 455
Median len: 300.0
Contigs > 1kb: 0
This is now our final set of targets that we will design baits from.
cd ..
mkdir finalAssembly
cd finalAssembly/
cp ../callSNPs/finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta ./
We'll run a few checks on this assembly to make sure things look good. First, confirm there are no N's:
grep -cP "^[ATCGN].*N" finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta
0
Next, we'll make sure that all of our targets here have full-length blast hits to original chimera-masked assembly:
blastn -db ../ARC/finished_allCTS/RBBHs.CTSonly6iter.chimeraMasked.fasta -query finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta -out finalTargets_bl2_RBBHsChimeraMasked.blast -outfmt 6
#!/usr/bin/perl
use strict;
use warnings;
use Bio::SearchIO;
use Bio::SeqIO;
my $in = shift;
my $seqs = shift;
my $out = shift;
my %seqHash;
my $seqIn = Bio::SeqIO->new(-file => $seqs,
-format => 'fasta');
while (my $seq = $seqIn->next_seq()) {
$seqHash{$seq->display_id()} = $seq->length();
}
my %blastHash;
my $searchIn = Bio::SearchIO->new(-file => $in,
-format => 'fasta');
while (my $result = $searchIn->next_result()) {
my $hit = $result->next_hit();
my $hsp = $result->next_hsp();
if ($hsp->frac_identical("total") < 1.0) {
die "Imperfect match for best HSP!\n";
}
$blastHash{$result->name()} = $hsp->length('query');
}
my $fullCounter = 0;
my $nonFullCounter = 0;
foreach my $target (sort keys %seqHash) {
if ($seqHash{$target} == $blastHash{$target}) {
$fullCounter++;
} else {
$nonFullCounter++;
}
}
print "Full-length matches: $fullCounter\n";
print "Incomplete matches: $nonFullCounter\n";perl checkForFullLengthMatches.pl finalTargets_bl2_RBBHsChimeraMasked.blast finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta
Full-length matches: 5236
Incomplete matches: 0
That looks good--we have 5,236 target sequences, and 5,236 full-length matches. We were only looking at the highest-scoring HSPs, and we made a check to make sure that the HSPs had 1.0 for a fraction of conserved bases in the match.
We'll once again visualize coverage across these target regions for the combined CTS and combined F1 reads:
mkdir mapping; mkdir mapping/depths
perl mapToFinalTargets.pl > ../logs/mapToFinalTargets.log 2>&1
cd mapping/depths
perl createF1andCTSdepthFiles.pl --out F1andCTSlocusDepthFiles
cd F1andCTSlocusDepthFile
mkdir plots
ls -al | grep "\.depth" | awk '{print $(NF)}' > depthFiles.txt
library("data.table")
depthFiles <- fread("depthFiles.txt", sep="\n", header=FALSE)
colors <- c("green", "blue")
# Now depthFiles$V1 holds all of the individual loci files--we'll read them one-by-one and create the plots
for (i in 1:length(depthFiles$V1)) {
dataz <- fread(depthFiles$V1[i])
samples <- unique(dataz$V1)
imageName <- paste("plots/", depthFiles$V1[i], ".png", sep="")
paste("Processing ", depthFiles$V1[i], ". locus: ", depthFiles$V1[i], "\n")
png(filename = imageName, width=1000, height=600, units="px", pointsize=12)
# Make the initial plot
plot(dataz$V3[dataz$V1 == samples[1]], dataz$V4[dataz$V1 == samples[1]], type="l", xlim=c(0,max(dataz$V3)), ylim=c(0,500), col=colors[1], lwd=1.2, main=depthFiles$V1[i], xlab="Position in contig", ylab="Read depth")
# Add all the lines for the rest of the samples
for (i in 2:length(samples)) {
lines(dataz$V3[dataz$V1 == samples[i]], dataz$V4[dataz$V1 == samples[i]], type="l", col=colors[i], lwd=1.5)
}
dev.off()
rm(dataz)
gc()
}The target set finalAssembly/finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta contains 80 of the original OPA sequences. We wish to reincorporate E15A3, as it was interesting in the previous analyses. It did not make it to the final target set in this analysis because it was not selected in the reciprocal best blast hit stages.
After pulling the sequence from the original "contigs.fasta" assembly, we put it into a new fasta file called E15A3.fasta. Then:
makeblastdb -in E15A3.fasta -dbtype nucl
blastn -db E15A3.fasta -query ../targetsNoBd.fasta -outfmt 6
This shows that the portion of the assembled contig that corresponds to the original target sequence in bases 558 to 948.
We'll pull out that sequence with the following script:
#!/usr/bin/perl
use strict;
use warnings;
use Bio::SeqIO;
my $seq = shift;
my $seq2 = shift;
my $seqIn = Bio::SeqIO->new(-file => $seq,
-format => 'fasta');
my $seqOut = Bio::SeqIO->new(-file => ">$seq2",
-format => 'fasta');
while (my $seq = $seqIn->next_seq()) {
my $overlappingSeq = $seq;
$overlappingSeq->seq($seq->subseq(558,948));
$seqOut->write_seq($overlappingSeq);
}perl subsetE15A3.pl E15A3.fasta E15A3_shortened.fasta
get_fasta_lengths.py --input E15A3_shortened.fasta
Reads: 1
Bp: 391
Avg. len: 391.0
STDERR len: nan
Min. len: 391
Max. len: 391
Median len: 391.0
Contigs > 1kb: 0
That's the right length, let's just make sure we pulled out the right bit:
makeblastdb -in E15A3_shortened.fasta -dbtype nucl
blastn -db E15A3_shortened.fasta -query ../targets.fasta -outfmt 6
Which outputs:
contig317689|E15A3|OPA allCTS_:_contig317689|E15A3|OPA_:_Contig001 98.47 391 6 0 1 391 391 1 0.0 689
So that looks right--the entire original target sequence overlaps with this new sequence we subseq'd from the assembly. We'll append it to the final target list:
cat finalTargetSetDepth5_atleast1SNP_min300bp_noMultiHSP_noNs.fasta E15A3_shortened.fasta > targetsToOrder.fasta
Then just remove the "NODE_1_length_2436_cov_65.0351_ID_1" from the last sequence identifier. The sequence identifier for this target will be slightly different from the others, because the others had the original target name appended to the sequence during the reciprocal best blast hit analysis.
Final target set to order specs:
get_fasta_lengths.py --input targetsToOrder.fasta
Reads: 5,237
Bp: 1,691,142
Avg. len: 322.921901852
STDERR len: 0.500010140248
Min. len: 300
Max. len: 455
Median len: 300.0
Contigs > 1kb: 0
We also wanted to generate assemblies using 6 ARC iterations on all of the read data from the 3 individuals. Here is the ARC config file for that:
## Name=value pairs:
## reference: contains reference sequences in fasta format
## numcycles: maximum number of times to try remapping
## mapper: the mapper to use (blat/bowtie2)
## assembler: the assembler to use (newbler/spades)
## nprocs: number of cores to use
## format: fasta or fasta, all must be the same
## verbose: control mapping/assembly log generation (True/False)
## urt: For Newbler, enable use read tips mode (True/False)
## map_against_reads: On iteration 1, skip assembly, map against mapped reads (True/False)
## assemblytimeout: kill assemblies and discard targets if they take longer than N minutes
##
## Columns:
## Sample_ID:Sample_ID
## FileName: path for fasta/fasta file
## FileType: PE1, PE2, or SE
## FileFormat: fasta or fasta
# reference=/mnt/Data1/HSEM020/targets.fasta
# numcycles=6
# mapper=bowtie2
# assembler=spades
# nprocs=20
# format=fastq
# verbose=True
# urt=True
# map_against_reads=False
# assemblytimeout=180
# bowtie2_k=5
# rip=True
# cdna=False
# subsample=1
# maskrepeats=True
# workingdirectory=/home/evan/manderReads/
Sample_ID FileName FileType
all3 All3.Ns.un1.fastq PE1
all3 All3.Ns.un2.fastq PE2
all3 All3.Ns.combinedJoinedAndSingles_trimmed.fastq SE
After doing the assembly with all individuals, the mapping actually performed worse for all samples (not just CTS). So we'll use the CTS-only assembly for this exercise.