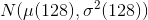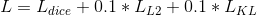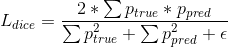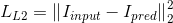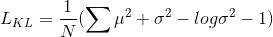This is an unofficial Pytorch implementation of BraTS_NvNet, based on Andriy Myronenko's paper on MICCAI_BraTS_2018_proceedings_shortPapers
BraTS2018: Multimodal Brain Tumor Segmentation Challenge 2018
The participants are called to address this task by using the provided clinically-acquired training data to develop their method and produce segmentation labels of the different glioma sub-regions. The sub-regions considered for evaluation are: 1) the "enhancing tumor" (ET), 2) the "tumor core" (TC), and 3) the "whole tumor" (WT).
Segmentation Labels:
- WT: 2
- TC: 1
- ET: 4
A combination of Vnet and VAE(variation auto-encoder).
Table 1. Encoder structure, where GN stands for group normalization(with group size of 8), Conv - 3*3*3 convolution, AddId - addition of identity / skip connection. Repeat column shows the number of repetitions of the block.
| Name | Ops | Repeat | Output size |
|---|---|---|---|
| Input | 4*160*192*128 | ||
| InitConv | Conv | 1 | 32*160*192*128 |
| EncoderBlock0 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 32*160*192*128 |
| EncoderDown1 | Conv stride 2 | 1 | 64*80*96*64 |
| EncoderBlock1 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 2 | 64*80*96*64 |
| EncoderDown2 | Conv stride 2 | 1 | 128*40*48*32 |
| EncoderBlock2 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 2 | 128*40*48*32 |
| EncoderDown3 | Conv stride 2 | 1 | 256*20*24*16 |
| EncoderBlock3 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 4 | 256*20*24*16 |
Table 2. Decoder structure, where Conv1 stands for 1*1*1 convolution, UpLinear - 3D linear spatial upsampling.
| Name | Ops | Repeat | Output size |
|---|---|---|---|
| DecoderUp2 | Conv1, UpLinear, +EncoderBlock2 | 1 | 128*40*48*32 |
| DecoderBlock2 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 128*40*48*32 |
| DecoderUp1 | Conv1, UpLinear, +EncoderBlock1 | 1 | 64*80*96*64 |
| DecoderBlock1 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 64*80*96*64 |
| DecoderUp0 | Conv1, UpLinear, +EncoderBlock0 | 1 | 32*160*192*128 |
| DecoderBlock0 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 32*160*192*128 |
| DecoderEnd | Conv1, Sigmoid | 1 | 1*160*192*128 |
Table 3. VAE decoder branch structure, where Dense stands for fully connected layer.
| Name | Ops | Repeat | Output size |
|---|---|---|---|
| VD | GN, ReLU, Conv (16) stride 2, Dense (256) | 1 | 256*1 |
| VDraw | sample | 1 | 128*1 |
| VU | Dense, ReLU, Conv1, UpLinear | 1 | 256*20*24*16 |
| VUp2 | Conv1, UpLinear | 1 | 128*40*48*32 |
| VBlock2 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 128*40*48*32 |
| VUp1 | Conv1, UpLinear | 1 | 64*80*96*64 |
| VBlock1 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 64*80*96*64 |
| VUp0 | Conv1, UpLinear | 1 | 32*160*192*128 |
| VBlock0 | GN, ReLU, Conv, GN, ReLU, Conv, AddId | 1 | 32*160*192*128 |
| Vend | Conv1 | 1 | 4*160*192*128 |
-
The loss function consists of 3 terms:
-
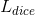 is applied to the decoder output
is applied to the decoder output 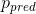 to match the segmentation mask
to match the segmentation mask 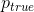 :
:where summation is voxel-wise, and the
 is a small constant to avoid zero division.
is a small constant to avoid zero division. -
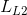 is appiled on the VAE branch output
is appiled on the VAE branch output 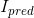 to match the input image
to match the input image 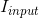 :
: -
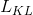 is standard VAE penalty term, a KL divergence between the estimated normal distribution
is standard VAE penalty term, a KL divergence between the estimated normal distribution 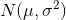 and a prior distribution
and a prior distribution 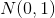 , which has a closed form representation:
, which has a closed form representation:where N is total number of image voxels.
-
The hyper-parameter weight of 0.1 was empirically set to provide a good balance between dice and VAE loss terms.
python: 3.6.2
pytorch: 0.4.1
pytables: 3.4.4
numpy: 1.13.1
tensorboardX: 1.4
nibabel: 2.3.0
nilearn: 0.4.2
tqdm: 4.26.0
pickle: 0.7.4
- data preprocessing:
- change the directory of brats 2018 training dataset and validation dataset
- set dataset format
- run the script:
python3 data_preprocess.py
- train model:
- set training data file path
- set training parameters
- run the script:
python3 main.py
- predict:
- set validation data file path
- set model file path
- run the script:
python3 predict.py
- make submission:
- set prediction file path
- set reconstruction parameters
- run the script:
python3 make_submission.py
- NvNet
- Loss
- DataLoader
- Train
- Predict
- Data Preprocess
- Make Submission