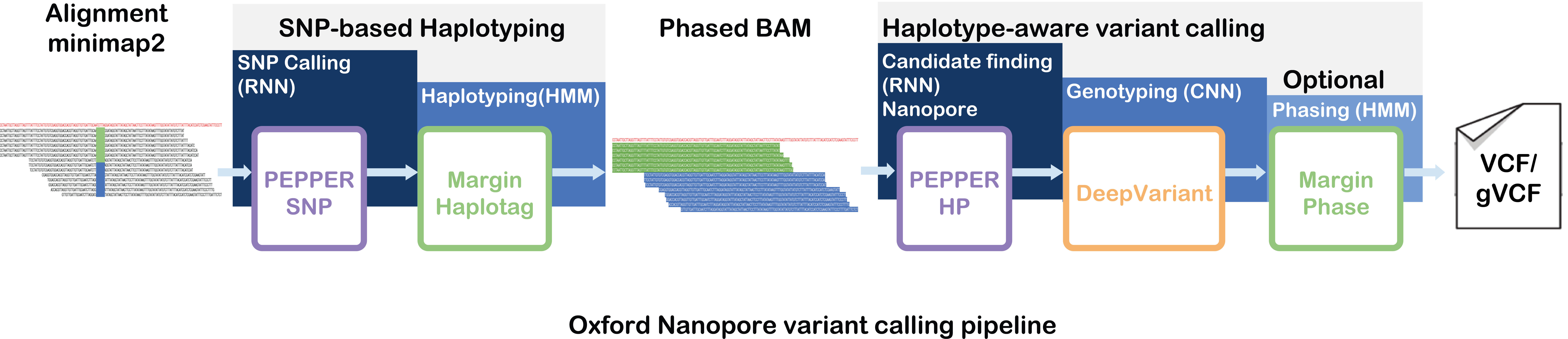
PEPPER is a genome inference module based on recurrent neural networks that enables long-read variant calling and nanopore assembly polishing in the PEPPER-Margin-DeepVariant pipeline. This pipeline enables nanopore-based variant calling with DeepVariant.
A detailed description of PEPPER-Margin-DeepVariant methods and results are discussed in the following manuscript:
bioRxiv: Haplotype-aware variant calling enables high accuracy in nanopore long-reads using deep neural networks.
Authors: Kishwar Shafin, Trevor Pesout, Pi-Chuan Chang, Maria Nattestad, Alexey Kolesnikov, Sidharth Goel,Gunjan Baid, Jordan M Eizenga, Karen H Miga, Paolo Carnevali, Miten Jain, Andrew Carroll, Benedict Paten.
Please follow the quickstart guides to assess your setup. Please follow case-study documentations for detailed instructions.
- Docker: Oxford Nanopore and PacBio HiFi variant calling quick start.
- Singularity: Oxford Nanopore and PacBio HiFi variant calling quick start.
The variant calling and assembly polishing pipelines can be run on Docker or Singularity. The case studies are designed on chr20 of HG002 sample.
Please pick the case-study of your pipeline of interest and the associated container runtime Docker or Singularity. The case-studies include input data and benchmarking of the run:
| Pipeline | Docker | Singularity | NVIDIA-docker (GPU) |
|---|---|---|---|
| Nanopore variant calling |
Link | Link | Link |
| PacBio HiFi variant calling |
Link | Link | Link |
| Nanopore assembly polishing with nanopore data |
Link | Link | Link |
| Nanopore assembly polishing with PacBio HiFi data |
Link | Link | Link |
- If you want to run
PEPPERorMarginindependent of the pipeline, please follow this documentation. - If you want to install
PEPPERlocally for development, please follow this documentation
PEPPER license, Margin License and DeepVariant License extend to the trained models (PEPPER, Margin and DeepVariant) and container environment (Docker and Singularity).
- Accuracy: Our pipeline won the precisionFDA truth challenge v2 for all benchmarking region and difficult to map region in the Oxford Nanopore category.
- Speed:
PEPPER-Margin-DeepVariantprovides a cheaper and faster solution to PacBio HiFi haplotype-aware variant calling. - Phased output:
PEPPER-Margin-DeepVariantcan produce high-quality phasing of variants without trio information with nanopore and PacBio HiFi reads.
We are thankful to the developers of these packages:
PEPPER-Margin-DeepVariant pipeline is developed in a collaboration between UC Santa Cruz genomics institute and the Genomics team in Google Health.
The name "P.E.P.P.E.R." is inspired from an A.I. created by Tony Stark in the Marvel Comics (Earth-616).
PEPPER is named after Tony Stark's then friend and the CEO of Resilient, Pepper Potts.