Interactive Multi-document Summarization Using Joint Optimization and Active Learning for Content Selection Grounded in User Feedback
In this project, we develop a general framework for Interactive Multi-Document Summarization. We propose an extractive multi-document summarization (MDS) system using joint optimization and active learning for content selection grounded in user feedback.
If you reuse this software, please use the following citation:
@inproceedings{TUD-CS-2017-0077,
title = {Joint Optimization of User-desired Content in Multi-document Summaries by Learning from User Feedback},
author = {P.V.S., Avinesh and Meyer, Christian M.},
publisher = {Association for Computational Linguistics},
volume = {Volume 1: Long Paper},
booktitle = {Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (ACL 2017)},
pages = {(to appear)},
month = aug,
year = {2017},
location = {Vancouver, Canada},
}
Abstract: In this paper, we propose an extractive multi-document summarization (MDS) system using joint optimization and active learning for content selection grounded in user feedback. Our method interactively obtains user feedback to gradually improve the results of a state-of-the-art integer linear programming (ILP) framework for MDS. Our methods complement fully automatic methods in producing high-quality summaries with a minimum number of iterations and feedbacks. We conduct multiple simulation-based experiments and analyze the effect of feedback-based concept selection in the ILP setup in order to maximize the user-desired content in the summary.
Contact person: Avinesh P.V.S., avinesh@aiphes.tu-darmstadt.de
http://www.ukp.tu-darmstadt.de/
Don't hesitate to send us an e-mail or report an issue, if something is broken (and it shouldn't be) or if you have further questions.
This repository contains experimental software and is published for the sole purpose of giving additional background details on the respective publication.
- python >= 2.7 (tested with 2.7.6)
-
Download ROUGE package from the link and place it in the rouge directory
>> mv RELEASE-1.5.5 rouge/ -
Install required python packages.
>> pip install -r requirements.txt -
Download the Standford Parser models and jars from the link
>> mv englishPCFG.ser.gz germanPCFG.ser.gz jars/ >> mv stanford-parser.jar stanford-parser-3.6.0-models.jar jars/ -
[Optional] To run the system for active learning models
Download the Google embeddings (English) from the [link](https://drive.google.com/file/d/0B7XkCwpI5KDYNlNUTTlSS21pQmM/) >> mkdir -p summarizer/data/embeddings/english >> mv GoogleNews-vectors-negative300.bin.gz summarizer/data/embeddings/english Download the News, Wikipedia embeddings (German) from the [link](https://public.ukp.informatik.tu-darmstadt.de/reimers/2014_german_embeddings/2014_tudarmstadt_german_50mincount.vec) >> mkdir -p summarizer/data/embeddings/german >> mv 2014_tudarmstadt_german_50mincount.vec summarizer/data/embeddings/german
-
Make sure that you have the raw datasets available. Each raw dataset needs to be extracted and follow the following directory structure:
+DUC_TEST | +--+docs | | | +-+d3103t | | | +-+ many files | | | +-+d31001t | +--+models | | | +-+ many files | +--+topics.xml -
Before running the pipeline, you have to preprocess the raw datasets using the
make_data.pyscript. Replace the DUC_TEST with appropriate dataset and run the same command.python summarizer/data_processer/make_data.py -d DUC_TEST -p summarizer/data/raw -a parse -l englishThe results should then be copied into a directory. We recommend using the
--iobasedirargument to set the directory+--+datasets/ | | | +--+raw/ | | | +--+DUC_TEST/ | | | | | +--+d31013t/ | | | | | +--+docs/ | | | | | +--+models/ | | | | +--+processed/ | | | +--+DUC_TEST/ | | | | | +--+d31013t/ | | | | | | | +--+docs/ | | | | | | | +--+docs.parsed/ | | | | | | | +--+summaries/ | | | | | | | +--+summaries.parsed/ | | | | | | | +--+summaries.upperbound/ | | | | | | | +--+task.json | | | | | +--+... | | | +--+ ... | +--+embeddings/ | +--+english/ | | | +-+GoogleNews-vectors-negative300.bin | | | +-+data/ | +--+german/ | +--+2014_tudarmstadt_german_50mincount.vec -
python pipeline.py --help for more details
python pipeline.py --summary_size=100 --oracle_type=accept_reject --data_set=DUC_TEST --summarizer_type=feedback pyhton pipeline.py --summary_size=100 --oracle_type=accept_reject --data_set=DUC_TEST --summarizer_type=baselines --language=english --rouge=rouge/RELEASE-1.5.5/ --iobasedir=outputs/
-
In DUC2004, task 5, topic d151h, document
APW20000104.0268produces andxml.etree.ElementTree.ParseError: mismatched tag: line 78, column 2The reason is a missing opening tag
<P>in row 72. -
In DUC2006, topic D0614E, Model Summary B
D0614.M.250.E.B. To fix it,Chrétienhas to be replaced byChretien. (Two times)
Verified by one (1) user.
-
download + install anaconda2 python 2.7.12 64bit from https://www.continuum.io/downloads#windows , e.g. https://repo.continuum.io/archive/Anaconda2-4.2.0-Windows-x86_64.exe
- take care that it is NOT python 2.7.13, as that version contains a regression bug which breaks pulp
TypeError: LoadLibrary() argument 1 must be string, not unicode -
download + install strawberry perl 64bit. In my case, Strawberry Perl (5.24.0.1-64bit).
-
download + install eclipse neon.2
-
download + instlal eclipse pydev
-
install perl module
XML::DOM -
install python modules
pip install -r requirements.txt
-
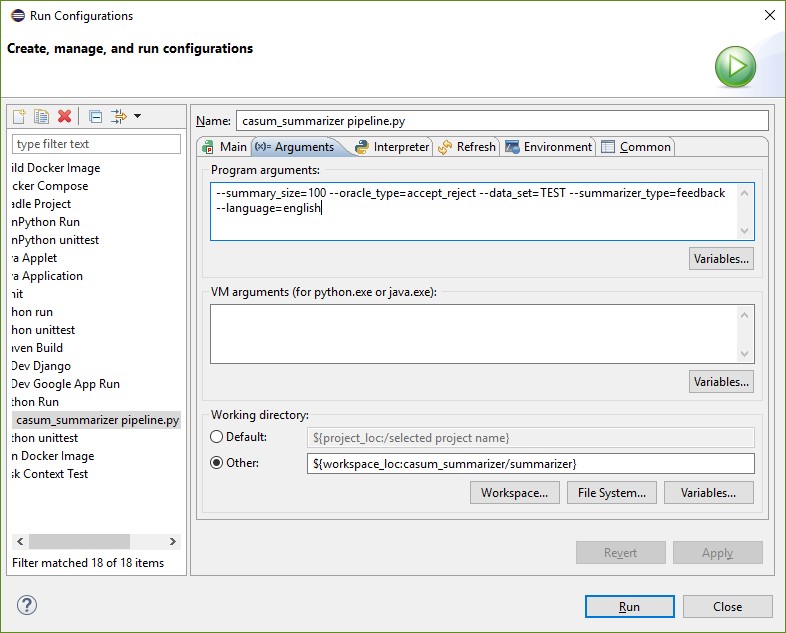
configure eclipse pydev run configuration as set up here:
--summary_size=100 --oracle_type=accept_reject --data_set=TEST --summarizer_type=feedback --language=english
-
Create a directory "tmp" on your root, e.g. "C:\tmp"!