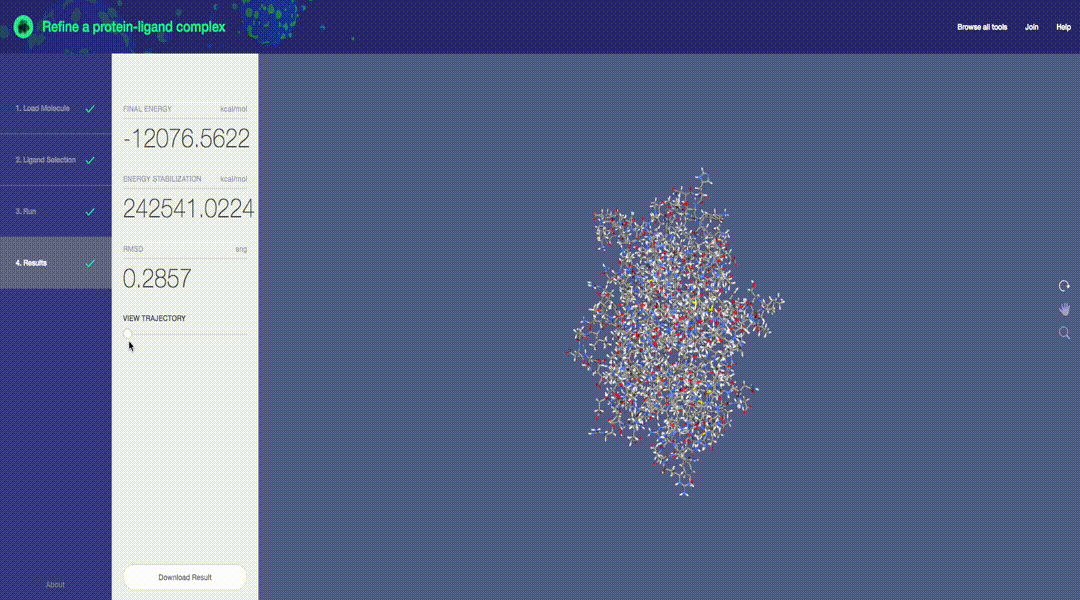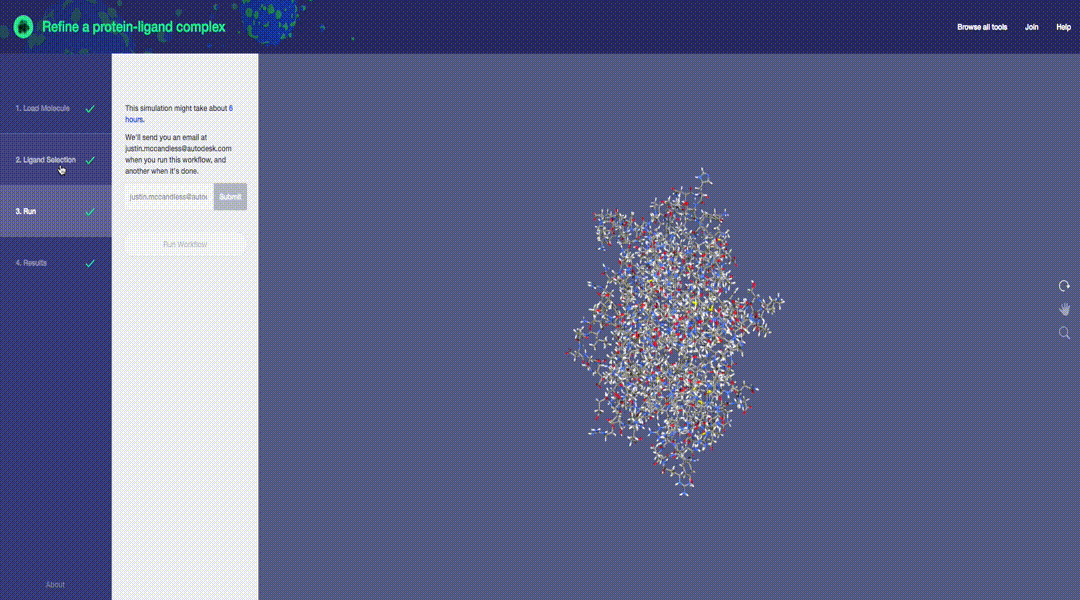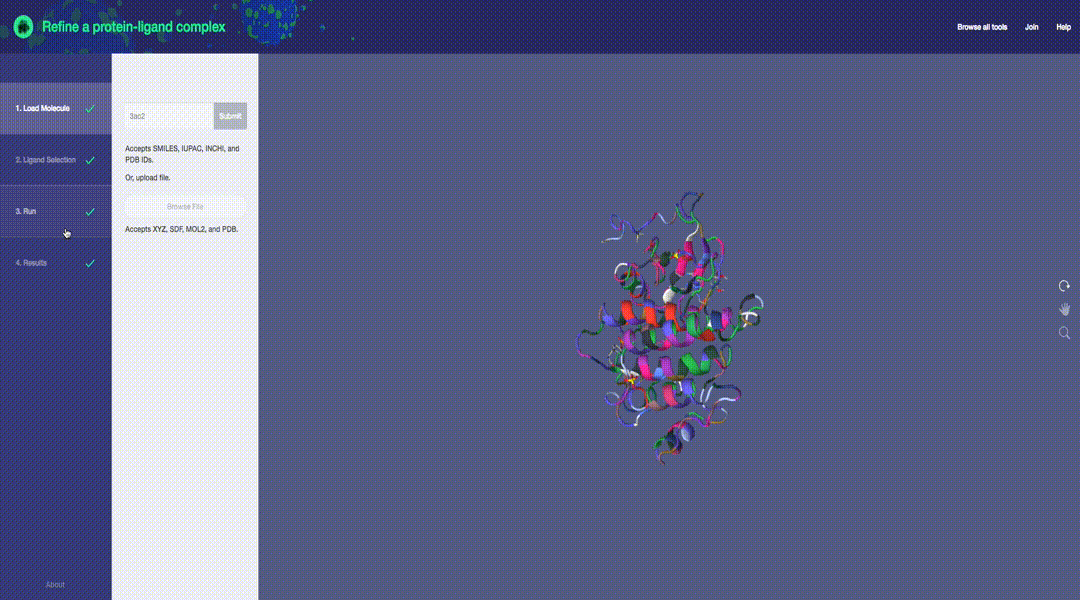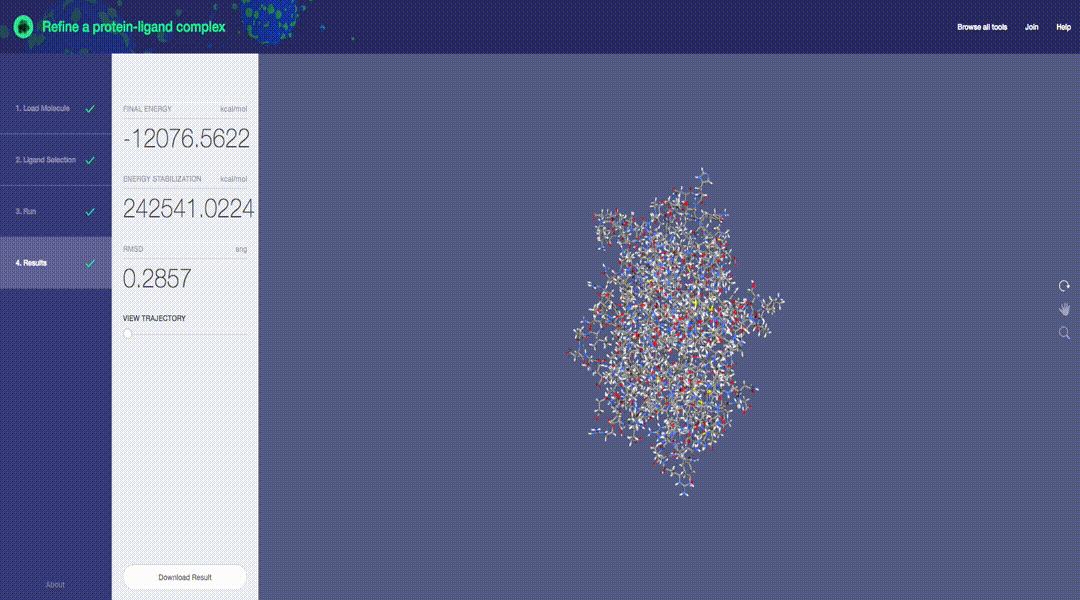
Predictive molecular modeling applications based on the Molecular Design Toolkit framework. (Early development, all features are subject to change)
git clone https://github.com/Autodesk/molecular-simulation-tools
cd molecular-design-applications
git submodule update --init --recursive
echo "0.0.1-local" > VERSION
docker-compose up
Then open your browser to http://localhost:4000
See client/README.md
npm run start
PORT: Sets the port that the server will run on.
SEND_GRID_API_KEY: Allows the server to send email via SendGrid.
URL: The url that the server can be accessed on publicly
FRONTEND_URL: The url root that we use for links in emails
GA_VIEW_ID: Used to retrieve number of views for each workflow from Google Analytics
Your Google Analytics key file should be placed at server/google_api_key.json. This will be used to fetch the view count from the Google Analytics API.
All routes are prefixed with the current version. See the mock server in client/test/fixtures/mock_server.js for example responses.
Begins an app session.
POST data:
{
"email": "some-user@gmail.com",
}
The email is a proxy for authentication.
Returns:
{
sessionId: <SessionID>
}
Update an app session outputs
POST data:
{
"widgetId1":
{
"outputPipeId1": {
"type": "inline",
"value": "the data"
},
"outputPipeId2": {
"type": "url",
"value": "http://some.data.url"
}
},
"widgetId2":
{
"outputPipeId3": {
"type": "inline",
"value": "the data"
},
"outputPipeId4": {
"type": "url",
"value": "http://some.data.url"
}
}
}
Returns GET /session/:sessionId
Update an app session outputs for a single widget
POST data:
{
"outputPipeId1": {
"type": "inline",
"encoding": "base64",
"value": "the data"
},
"outputPipeId2": {
"type": "url",
"value": "http://some.data.url"
}
}
Returns GET /session/:sessionId
Delete widget outputs
POST data:
[
"widgetId1",
"widgetId2"
]
Returns GET /session/:sessionId
Returns the session state
{
"session": ":sessionId",
"widgets": {
"widgetId1": {
"out": {
"outputPipe1": {
"type": "inline",
"value": "actual data string",
"encoding": "utf8"
},
"outputPipe2": {
"type": "url",
"value": "http://url.to.data"
}
}
}
}
}
Submits a "fast" docker job to the server. Turbo jobs are not saved, and there is no interaction with sessions or widgets. It is meant for client widgets to call on their own.
POST data (all fields are optional):
{
"id": "optional custom job id",
"inputs": [
{
"name": "input1Key",
"type": "inline",
"value": "input1ValueString",
"encoding": "base64"
},
{
"name": "input2Key",
"type": "url",
"value": "http://some.url.value"
}
],
"image": "docker.io/busybox:latest",
"imagePullOptions": {},
"command": ["/bin/sh", "/some/script"],
"workingDir": "/inputs",
"parameters": {
"cpus": 1,
"maxDuration": 600
},
"inputsPath": "/inputs",
"outputsPath": "/ouputs",
"meta": {}
}
Returns:
{
"id": "S1DWbFHTx",
"outputs": [
{
"name": "val1",
"value": "Some string data",
"encoding": "utf8"
},
{
"name": "val2",
"value": "20731535302965964",
"encoding": "base64"
}
],
"error": null,
"stdout": [],
"stderr": [],
"exitCode": 0,
"stats": {
"copyInputs": "0.149s",
"ensureImage": "0s",
"containerCreation": "0.161s",
"containerExecution": "1.302s",
"copyOutputs": "0.208s",
"copyLogs": "0.003s",
"total": "1.5110000000000001s"
}
}
Submits a CCC job for a session widget. The outputs will be saved in the session and the session will be updated (and notified via websocket)
POST data (all fields optional):
{
"inputs": {
"inputKey1": {
"type": "url",
"value": "http://some.url"
},
"inputKey2": {
"type": "inline",
"value": "Raw data string"
},
},
"image": "docker.io/busybox:latest",
"imagePullOptions": {},
"command": ["/bin/sh", "/some/script"],
"workingDir": "/inputs",
"parameters": {
"cpus": 1,
"maxDuration": 600
},
"inputsPath": "/inputs",
"outputsPath": "/ouputs",
"meta": {},
"appendStdOut": true,
"appendStdErr": true
}
Returns:
{
"sessionId": "fefd7b397aaf4a7c8ce2a6d8fbd28759",
"jobId": "BJaHVYBTl"
}
The results will be computed out-of-band, and returned via the websocket (the server monitors the job internally).
A websocket request to the app root will initiate a websocket connection
After the connection is established, the server needs to know what the app session is, by the client sending a message (stringified):
{
"jsonrpc": "2.0",
"method": jsonrpcConstants.SESSION,
"params": {
"sessionId": "<session/run id>"
}
}
The server will then send the latest session data, and will send a new state any time the state is changed on the server:
{
"session": "<session/runId>",
"widgets": {
"widget1": {
"out": {
"widget1pipe1": {
"type": "inline",
"value": "widgetId1Pipe1Value"
}
}
},
"widget2": {
"out": {
"widget2pipe1": {
"type": "inline",
"value": "widgetId2Pipe1Value"
}
}
}
}
}
When the app session is changed, the websocket will automatically close.
Returns the indicated workflow.
Returns the stdout logs of the finished workflow
Returns the stderr logs of the finished workflow
Returns the exit code of the finished workflow
Returns the indicated run with its workflow populated.
Runs the indicated workflow. Requires workflowId, email, and pdbUrl.
Returns the pdb data and a url to the pdb file represented by the given pdbId. For some workflow ids, the pdb will be processed before being returned, and a data parameter will also be included with additional data.
Uploads the given pdb file to the server and returns a public URL to it. Sends formdata with a workflowId and a file.
These methods are not prefixed with the version tag. I.e. they are e.g. http://localhost:4000/test and not http://localhost:4000/v1/test.
Checks connectivity to the CCC compute server. Does not test workflows.
Tests the workflow with baked in test data
Tests all workflows
For development:
curl --max-time 10000 localhost:4000/test/all
Currently, Redis needs to be seeded with at least one workflow for the app to use, which you can create with:
hset workflows 0 '{"id": 0, "title": "VDE"}'
In addition to mounting local directories as mentioned above, you can recompile the frontend assets on change by running npm run watch in the client directory.
This project is developed and maintained by the Molecular Design Toolkit project. Please see that project's CONTRIBUTING document for details.
Copyright 2016 Autodesk Inc.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
We use Autodesk Molecule Viewer to display and navigate molecular data. Autodesk Molecule Viewer is not released under an open source license. For more information about the Autodesk Molecule Viewer license please refer to: https://molviewer.com/molviewer/docs/Pre-Release_Product_Testing_Agreement.pdf.