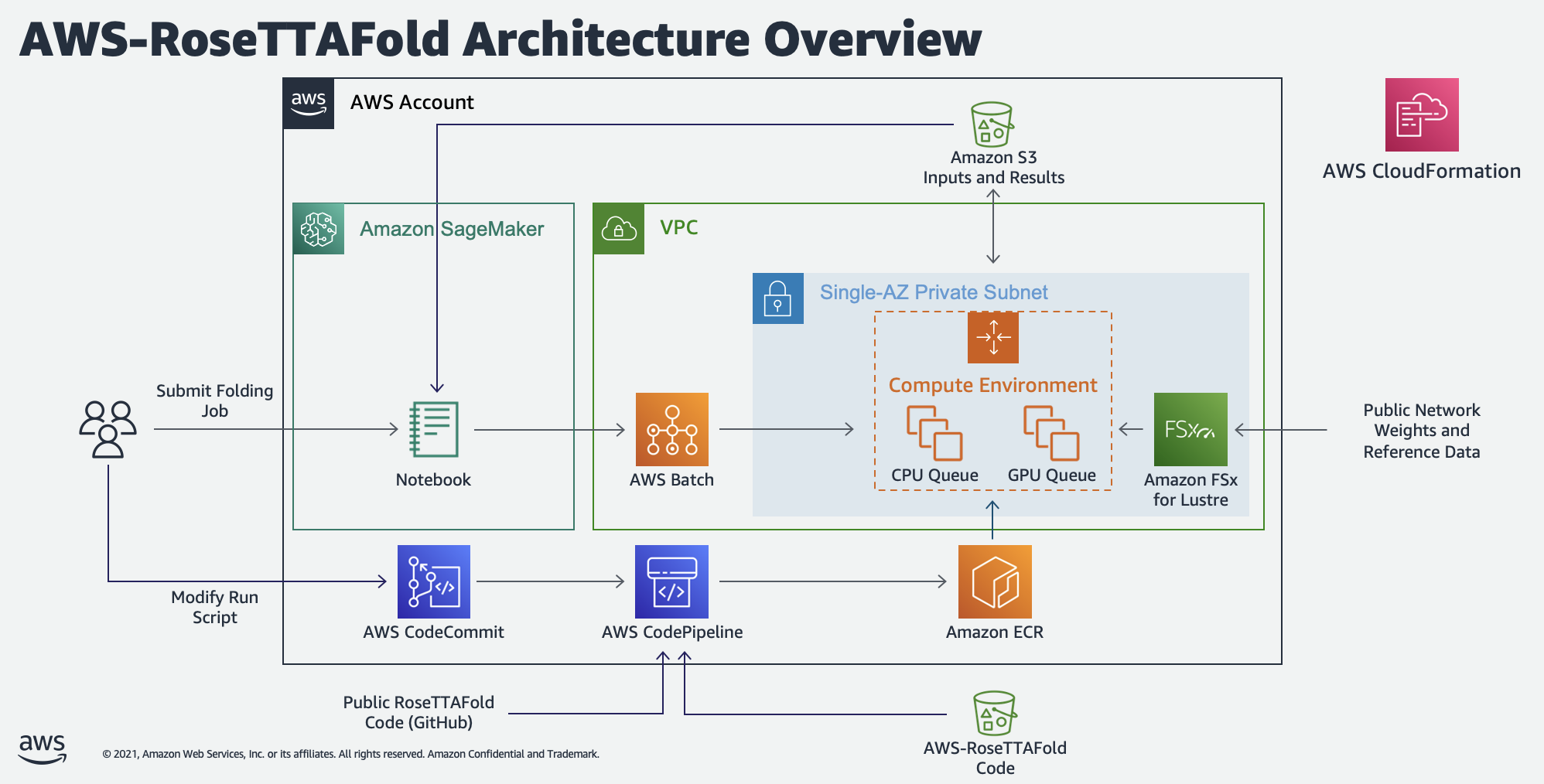
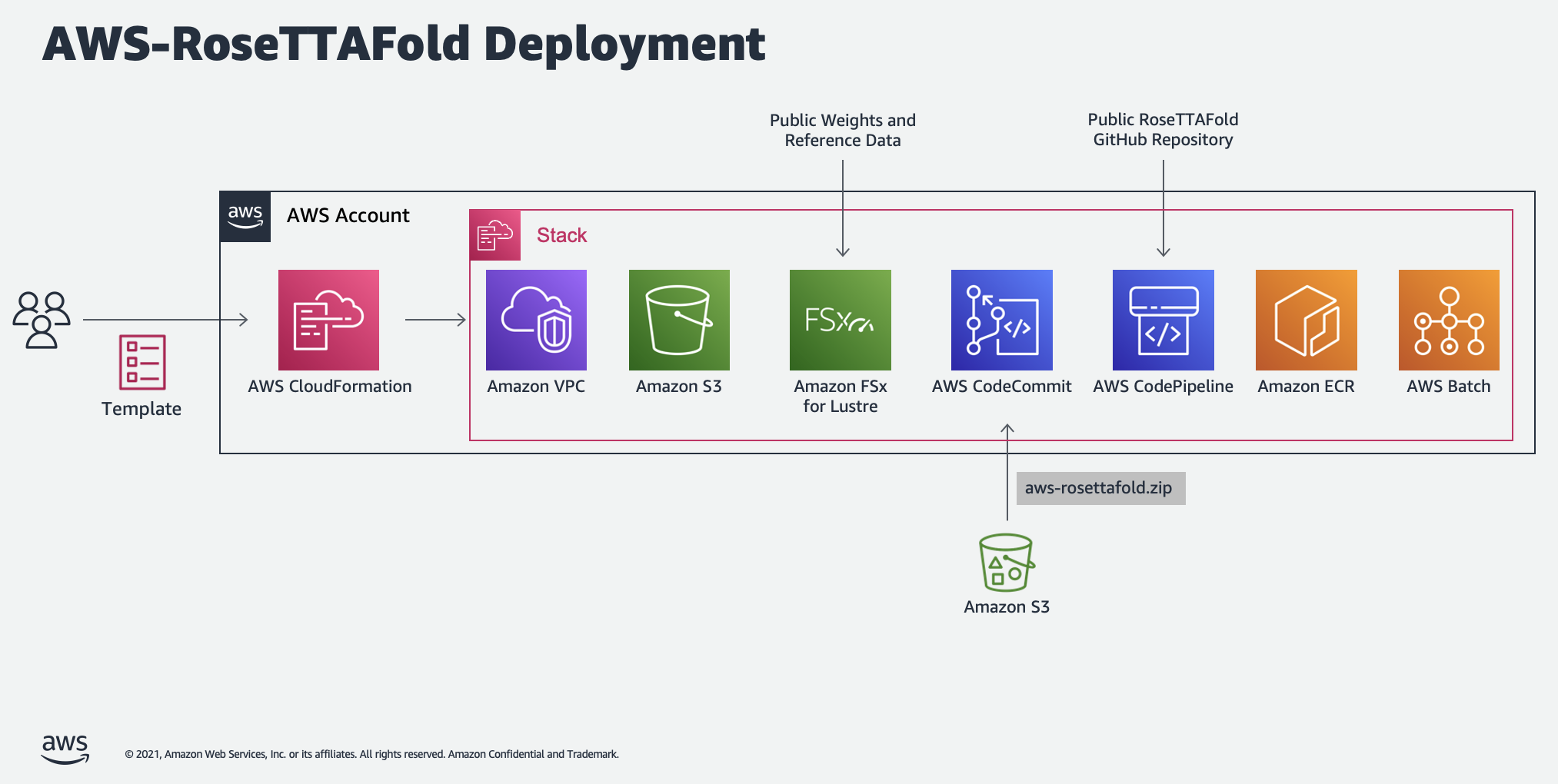
Infrastructure template and Jupyter notebooks for running RoseTTAFold on AWS Batch.
PProteins are large biomolecules that play an important role in the body. Knowing the physical structure of proteins is key to understanding their function. However, it can be difficult and expensive to determine the structure of many proteins experimentally. One alternative is to predict these structures using machine learning algorithms. Several high-profile research teams have released such algorithms, including AlphaFold 2, RoseTTAFold, and others. Their work was important enough for Science magazine to name it the "2021 Breakthrough of the Year".
Both AlphaFold 2 and RoseTTAFold use a multi-track transformer architecture trained on known protein templates to predict the structure of unknown peptide sequences. These predictions are heavily GPU-dependent and take anywhere from minutes to days to complete. The input features for these predictions include multiple sequence alignment (MSA) data. MSA algorithms are CPU-dependent and can themselves require several hours of processing time.
Running both the MSA and structure prediction steps in the same computing environment can be cost inefficient, because the expensive GPU resources required for the prediction sit unused while the MSA step runs. Instead, using a high performance computing (HPC) service like AWS Batch (https://aws.amazon.com/batch/) allows us to run each step as a containerized job with the best fit of CPU, memory, and GPU resources.
In this post, we demonstrate how to provision and use AWS Batch and other services to run AI-driven protein folding algorithms like RoseTTAFold.
-
Choose Launch Stack:
-
For Stack Name, enter a value unique to your account and region.
-
For StackAvailabilityZone choose an availability zone.
-
Select I acknowledge that AWS CloudFormation might create IAM resources with custom names.
-
Choose Create stack.
-
Wait approximately 30 minutes for AWS CloudFormation to create the infrastructure stack and AWS CodeBuild to build and publish the AWS-RoseTTAFold container to Amazon Elastic Container Registry (Amazon ECR).
Option 1: Mount the FSx for Lustre file system to an EC2 instance
- Sign in to the AWS Management Console and open the Amazon EC2 console at https://console.aws.amazon.com/ec2.
- In the navigation pane, under Instances, select Launch Templates.
- Choose the Launch template ID for your stack, such as
aws-rosettafold-launch-template-stack-id-suffix. - Choose Actions, Launch instance from template.
- Launch a new EC2 instance and connect using either SSH or SSM.
- Download and extract the network weights and sequence database files to the attached volume at
/fsx/aws-rosettafold-ref-dataaccording to installation steps 3 and 5 from the RoseTTAFold public repository.
Option 2: Lazy-load the data from a S3 data repository
- Create a new S3 bucket in your region of interest.
- Download and extract the network weights and sequence database files as described above and transfer them to your S3 bucket.
- Sign in to the AWS Management Console and open the Amazon FSx for Lustre console at https://console.aws.amazon.com/fsx.
- Choose the File System name for your stack, such as
aws-rosettafold-fsx-lustre-stack-id-suffix. - On the file system details page, choose Data repository, Create data repository association.
- For File system path enter
/aws-rosettafold-ref-data. - For Data repository path enter the s3 url for your new S3 bucket.
- Choose Create.
Creating the data repository association will immediately load the file metadata to the file system. However, the data itself will not be available until requested by a job. This will add several hours to the duration of the first job you submit. However, subsequent jobs will complete much faster.
Once you have finished loading the model weights and sequence data base files, the FSx for Lustre file system will include the following files:
/fsx
└── /aws-rosettafold-ref-data
├── /bfd
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_a3m.ffdata (1.4 TB)
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_a3m.ffindex (1.7 GB)
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_cs219.ffdata (15.7 GB)
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_cs219.ffindex (1.6 GB)
│ ├── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_hhm.ffdata (304.4 GB)
│ └── bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt_hhm.ffindex (123.6 MB)
├── /pdb100_2021Mar03
│ ├── LICENSE (20.4 KB)
│ ├── pdb100_2021Mar03_a3m.ffdata (633.9 GB)
│ ├── pdb100_2021Mar03_a3m.ffindex (3.9 MB)
│ ├── pdb100_2021Mar03_cs219.ffdata (41.8 MB)
│ ├── pdb100_2021Mar03_cs219.ffindex (2.8 MB)
│ ├── pdb100_2021Mar03_hhm.ffdata (6.8 GB)
│ ├── pdb100_2021Mar03_hhm.ffindex (3.4 GB)
│ ├── pdb100_2021Mar03_pdb.ffdata (26.2 GB)
│ └── pdb100_2021Mar03_pdb.ffindex (3.7 MB)
├── /UniRef30_2020_06
│ ├── UniRef30_2020_06_a3m.ffdata (139.6 GB)
│ ├── UniRef30_2020_06_a3m.ffindex (671.0 MG)
│ ├── UniRef30_2020_06_cs219.ffdata (6.0 GB)
│ ├── UniRef30_2020_06_cs219.ffindex (605.0 MB)
│ ├── UniRef30_2020_06_hhm.ffdata (34.1 GB)
│ ├── UniRef30_2020_06_hhm.ffindex (19.4 MB)
│ └── UniRef30_2020_06.md5sums (379.0 B)
└── /weights
├── RF2t.pt (126 MB KB)
├── Rosetta-DL_LICENSE.txt (3.1 KB)
├── RoseTTAFold_e2e.pt (533 MB)
└── RoseTTAFold_pyrosetta.pt (506 MB)
- Clone the CodeCommit repository created by CloudFormation to a Jupyter Notebook environment of your choice.
- Use the
AWS-RoseTTAFold.ipynbandCASP14-Analysis.ipynbnotebooks to submit protein sequences for analysis.
This project creates two computing environments in AWS Batch to run the "end-to-end" protein folding workflow in RoseTTAFold. The first of these uses the optimal mix of c4, m4, and r4 instance types based on the vCPU and memory requirements specified in the Batch job. The second environment uses g4dn on-demand instances to balance performance, availability, and cost.
A scientist can create structure prediction jobs using one of the two included Jupyter notebooks. AWS-RoseTTAFold.ipynb demonstrates how to submit a single analysis job and view the results. CASP14-Analysis.ipynb demonstrates how to submit multiple jobs at once using the CASP14 target list. In both of these cases, submitting a sequence for analysis creates two Batch jobs, one for data preparation (using the CPU computing environment) and a second, dependent job for structure prediction (using the GPU computing environment).
Both the data preparation and structure prediction use the same Docker image for execution. This image, based on the public Nvidia CUDA image for Ubuntu 20, includes the v1.1 release of the public RoseTTAFold repository, as well as additional scripts for integrating with AWS services. CodeBuild will automatically download this container definition and build the required image during stack creation. However, end users can make changes to this image by pushing to the CodeCommit repository included in the stack. For example, users could replace the included MSA algorithm (hhblits) with an alternative like MMseqs2 or replace the RoseTTAFold network with an alternative like AlphaFold 2 or Uni-Fold.
This workload costs approximately $760 per month to maintain, plus another $0.50 per job.
Running the CloudFormation template at config/cfn.yaml creates the following resources in the specified availability zone:
- A new VPC with a private subnet, public subnet, NAT gateway, internet gateway, elastic IP, route tables, and S3 gateway endpoint.
- A FSx Lustre file system with 1.2 TiB of storage and 1,200 MB/s throughput capacity. This file system can be linked to an S3 bucket for loading the required reference data when the first job executes.
- An EC2 launch template for mounting the FSX file system to Batch compute instances.
- A set of AWS Batch compute environments, job queues, and job definitions for running the CPU-dependent data prep job and a second for the GPU-dependent prediction job.
- CodeCommit, CodeBuild, CodePipeline, and ECR resources for building and publishing the Batch container image. When CloudFormation creates the CodeCommit repository, it populates it with a zipped version of this repository stored in a public S3 bucket. CodeBuild uses this repository as its source and adds additional code from release 1.1 of the public RoseTTAFold repository. CodeBuild then publishes the resulting container image to ECR, where Batch jobs can use it as needed.
This library is licensed under the MIT-0 License. See the LICENSE file for more information.
The University of Washington has made the code and data in the RoseTTAFold public repository available under an MIT license. However, the model weights used for prediction are only available for internal, non-profit, non-commercial research use. For information, please see the full license agreement and contact the University of Washington for details.
See CONTRIBUTING for more information.