SDK and CLI Tools for working with the AWS HealthOmics Service.
AWS HealthOmics Tools is available through pypi. To install, type:
pip install amazon-omics-toolsTo install from source:
git clone https://github.com/awslabs/amazon-omics-tools.git
pip install ./amazon-omics-tools
The TransferManager class makes it easy to download files from a AWS HealthOmics reference or read set. By default the files are saved to the current directory, or you can specify a custom location with the directory parameter.
import boto3
from omics.common.omics_file_types import ReadSetFileName, ReferenceFileName, ReadSetFileType
from omics.transfer.manager import TransferManager
from omics.transfer.config import TransferConfig
REFERENCE_STORE_ID = "<my-reference-store-id>"
SEQUENCE_STORE_ID = "<my-sequence-store-id>"
client = boto3.client("omics")
manager = TransferManager(client)
# Download all files for a reference.
manager.download_reference(REFERENCE_STORE_ID, "<my-reference-id>")
# Download all files for a read set to a custom directory.
manager.download_read_set(SEQUENCE_STORE_ID, "<my-read-set-id>", "my-sequence-data")Specific files can be downloaded via the download_reference_file and download_read_set_file methods.
The client_fileobj parameter can be either the name of a local file to create for storing the data, or a TextIO or BinaryIO object that supports write methods.
# Download a specific reference file.
manager.download_reference_file(
REFERENCE_STORE_ID,
"<my-reference-id>",
ReferenceFileName.INDEX
)
# Download a specific read set file with a custom filename.
manager.download_read_set_file(
SEQUENCE_STORE_ID,
"<my-read-set-id>",
ReadSetFileName.INDEX,
"my-sequence-data/read-set-index"
)Specific files can be uploaded via the upload_read_set method.
The fileobjs parameter can be either the name of a local file, or a TextIO or BinaryIO object that supports read methods.
For paired end reads, you can define fileobjs as a list of files.
# Upload a specific read set file.
read_set_id = manager.upload_read_set(
"my-sequence-data/read-set-file.bam",
SEQUENCE_STORE_ID,
"BAM",
"name",
"subject-id",
"sample-id",
"<my-reference-arn>",
)
# Upload paired end read set files.
read_set_id = manager.upload_read_set(
["my-sequence-data/read-set-file_1.fastq.gz", "my-sequence-data/read-set-file_2.fastq.gz"],
SEQUENCE_STORE_ID,
"FASTQ",
"name",
"subject-id",
"sample-id",
"<my-reference-arn>",
)Transfer events: on_queued, on_progress, and on_done can be observed by defining a subclass of OmicsTransferSubscriber and passing in an object which can receive events.
class ProgressReporter(OmicsTransferSubscriber):
def on_queued(self, **kwargs):
future: OmicsTransferFuture = kwargs["future"]
print(f"Download queued: {future.meta.call_args.fileobj}")
def on_done(self, **kwargs):
print("Download complete")
manager.download_read_set(SEQUENCE_STORE_ID, "<my-read-set-id>", subscribers=[ProgressReporter()])Transfer operations use threads to implement concurrency. Thread use can be disabled by setting the use_threads attribute to False.
If thread use is disabled, transfer concurrency does not occur. Accordingly, the value of the max_request_concurrency attribute is ignored.
# Disable thread use/transfer concurrency
config = TransferConfig(use_threads=False)
manager = TransferManager(client, config)
manager.download_read_set(SEQUENCE_STORE_ID, "<my-read-set-id>")The OmicsUriParser class makes it easy to parse AWS HealthOmics readset and reference URIs to extract fields relevant for calling
AWS HealthOmics APIs.
Readset file URIs come in the following format:
omics://<AWS_ACCOUNT_ID>.storage.<AWS_REGION>.amazonaws.com/<SEQUENCE_STORE_ID>/readSet/<READSET_ID>/<SOURCE1/SOURCE2>
For example:
omics://123412341234.storage.us-east-1.amazonaws.com/5432154321/readSet/5346184667/source1
omics://123412341234.storage.us-east-1.amazonaws.com/5432154321/readSet/5346184667/source2
Reference file URIs come in the following format:
omics://<AWS_ACCOUNT_ID>.storage.<AWS_REGION>.amazonaws.com/<REFERENCE_STORE_ID>/reference/<REFERENCE_ID>/source
For example:
omics://123412341234.storage.us-east-1.amazonaws.com/5432154321/reference/5346184667/source
To handle both HealthOmics URI types, you would use code like the following:
import boto3
from omics.uriparse.uri_parse import OmicsUriParser, OmicsUri
READSET_URI_STRING = "omics://123412341234.storage.us-east-1.amazonaws.com/5432154321/readSet/5346184667/source1"
REFERENCE_URI_STRING = "omics://123412341234.storage.us-east-1.amazonaws.com/5432154321/reference/5346184667/source"
client = boto3.client("omics")
readset = OmicsUriParser(READSET_URI_STRING).parse()
reference = OmicsUriParser(REFERENCE_URI_STRING).parse()
# use the parsed fields from the URIs to call AWS HealthOmics APIs:
manager = TransferManager(client)
# Download all files for a reference.
manager.download_reference(reference.store_id, reference.resource_id)
# Download all files for a read set to a custom directory.
manager.download_read_set(readset.store_id, readset.resource_id, readset.file_name)
# Download a specific read set file with a custom filename.
manager.download_read_set_file(
readset.store_id,
readset.resource_id,
readset.file_name,
"my-sequence-data/read-set-index"
)CLI tools are modules in this package that can be invoked from the command line with:
python -m omics.cli.<TOOL-NAME>The omics-rerun tool makes it easy to start a new run execution from a CloudWatch Logs manifest.
For an overview of what it does and available options run:
python -m omics.cli.rerun -hThe following example lists all workflow run ids which were completed on July 1st (UTC time):
python -m omics.cli.rerun -s 2023-07-01T00:00:00 -e 2023-07-02T00:00:00this returns something like:
1234567 (2023-07-01T12:00:00.000)
2345678 (2023-07-01T13:00:00.000)
To rerun a previously-executed run, specify the run id you would like to rerun:
python -m omics.cli.rerun 1234567this returns something like:
StartRun request:
{
"workflowId": "4974161",
"workflowType": "READY2RUN",
"roleArn": "arn:aws:iam::123412341234:role/MyRole",
"parameters": {
"inputFASTQ_2": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R2_001-5x.fastq.gz",
"inputFASTQ_1": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R1_001-5x.fastq.gz"
},
"outputUri": "s3://my-bucket/my-path"
}
StartRun response:
{
"arn": "arn:aws:omics:us-west-2:123412341234:run/3456789",
"id": "3456789",
"status": "PENDING",
"tags": {}
}
It is possible to override a request parameter from the original run. The following example tags the new run, which is particularly useful as tags are not propagated from the original run.
python -m omics.cli.rerun 1234567 --tag=myKey=myValue
this returns something like:
StartRun request:
{
"workflowId": "4974161",
"workflowType": "READY2RUN",
"roleArn": "arn:aws:iam::123412341234:role/MyRole",
"parameters": {
"inputFASTQ_2": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R2_001-5x.fastq.gz",
"inputFASTQ_1": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R1_001-5x.fastq.gz"
},
"outputUri": "s3://my-bucket/my-path",
"tags": {
"myKey": "myValue"
}
}
StartRun response:
{
"arn": "arn:aws:omics:us-west-2:123412341234:run/4567890",
"id": "4567890",
"status": "PENDING",
"tags": {
"myKey": "myValue"
}
}
Before submitting a rerun request, it is possible to dry-run to view the new StartRun request:
python -m omics.cli.rerun -d 1234567this returns something like:
StartRun request:
{
"workflowId": "4974161",
"workflowType": "READY2RUN",
"roleArn": "arn:aws:iam::123412341234:role/MyRole",
"parameters": {
"inputFASTQ_2": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R2_001-5x.fastq.gz",
"inputFASTQ_1": "s3://omics-us-west-2/sample-inputs/4974161/HG002-NA24385-pFDA_S2_L002_R1_001-5x.fastq.gz"
},
"outputUri": "s3://my-bucket/my-path"
}
The omics-run-analyzer tool retrieves a workflow run manifest from CloudWatchLogs and generates statistics for the run, including CPU and memory utilization for each workflow task.
For an overview of what it does and available options run:
python -m omics.cli.run_analyzer -hThe following example lists all workflow runs completed in the past 5 days:
python -m omics.cli.run_analyzer -t5dthis returns something like:
Workflow run IDs (<completionTime> <UUID>):
1234567 (2024-02-01T12:00:00 12345678-1234-5678-9abc-123456789012)
2345678 (2024-02-03T13:00:00 12345678-1234-5678-9abc-123456789012)
python -m omics.cli.run_analyzer 1234567 -o run-1234567.csvthis returns something like:
omics-run-analyzer: wrote run-1234567.csv
The CSV output by the command above includes the following columns:
- arn : Unique workflow run or task identifier
- type : Resource type (run or task)
- name : Workflow run or task name
- startTime : Start timestamp for the workflow run or task (UTC time)
- stopTime : Stop timestamp for the workflow run or task (UTC time)
- runningSeconds : Approximate workflow run or task runtime (in seconds)
- cpusRequested : The number of vCPU requested by the workflow task
- gpusRequested : The number of GPUs requested by the workflow task
- memoryRequestedGiB : Gibibytes of memory requested by the workflow task
- omicsInstanceTypeReserved : Requested HealthOmics instance type for each task
- omicsInstanceTypeMinimum : Minimum HealthOmics instance type that could run each task.
- recommendedCpus: The number of CPUs recommended for this task (corresponding to the number of CPUs in the omicsInstanceTypeMinimum)
- recommendedMemoryGiB: The amount of GiB of memory recommended for this task (corresponding to the number of CPUs in the omicsInstanceTypeMinimum)
- estimatedUSD : Estimated HealthOmics charges (USD) for the workflow based on sizeReserved and runningSeconds
- minimumUSD : Estimated HealthOmics charges (USD) for the workflow based on the recommended omicsInstanceTypeMinimum and runningSeconds
- cpuUtilizationRatio : CPU utilization (cpusMaximum / cpusReserved) for workflow task(s)
- memoryUtilizationRatio : Memory utilization (memoryMaximumGiB / memoryReservedGiB) for the workflow task(s)
- storageUtilizationRatio : Storage utilization (storageMaximumGiB / storageReservedGiB) for the workflow run
- cpusReserved : vCPUs reserved for workflow task(s)
- cpusMaximum : Maximum vCPUs used during a single 1-minute interval
- cpusAverage : Average vCPUs used by workflow task(s)
- gpusReserved : GPUs reserved for workflow task(s)
- memoryReservedGiB : Gibibytes of memory reserved for workflow task(s)
- memoryMaximumGiB : Maximum gibibytes of memory used during a single 1-minute interval
- memoryAverageGiB : Average gibibytes of memory used by workflow task(s)
- storageReservedGiB : Gibibytes of storage reserved for the workflow run
- storageMaximumGiB : Maximum gibibytes of storage used during a single 1-minute interval
- storageAverageGiB : Average gibibytes of storage used by the workflow run
For rows that are a task type, the maximums, averages and reserved columns refer to the maximum, average or reserved amounts of the respective resource used by that task. These values can be used to guide the resources that should be requested for that task. For rows that are a run type the maximums, averages and reserved columns refer to the maximum, average or reserved amounts of the respective resource used concurrently by that run. These values can be used to determine if the accounts HealthOmics active CPUs/memory limits are being reached which might indicate the run is constrained by these limits.
Warning
At this time AWS HealthOmics does not report the average or maximum storage used by runs that use "DYNAMIC" storage that run for under two hours. Because of this limitation the storageMaximumGiB and storageAverageGiB are set to zero and will not be included in the estimate run cost.
Based on the metrics observed and calculated for a run, the application will recommend the smallest instance type that could be used for each task in the run. The type is reported in the omicsInstanceTypeMinimum column. To obtain this type for a task you can set the task CPU and memory requested for the task to the values of recommendedCpus and recommendedMemoryGiB in you workflow definition. Based on this change each task would be estimated to
reduce the cost of the run by estimatedUSD minus minimumUSD. The total potential cost reduction for the entire run can be estimated by subtracting the minimumUSD value from the estimatedUSD value in the row where the type is "run".
Sometimes you will see variance in the amount of memory and CPU used in a run task, especially if you expect to run workflows with larger input files than were used in the analyzed run. For this reason you might want to allow add some headroom to the recommendations produced by the the run analyzer.
The -H or --headroom flag can be use to add an additional 0.0 to 1.0 times the max CPU or memory used by a task to the calculation used to determine
the omicsInstanceTypeMinimum recommendation. For example if a task used a max of 3.6 GiB of memory and the headroom value is 0.5 then 6 GiB of memory - math.ceil(3.6 * (1 + 0.5)) - will be used to determine the minimum instance type that should be used.
If your analyzed run is already close to optimal then adding headroom might result in the recommended minimum instance being larger than the instance used in the run which will also cause the "minimumUSD" to be larger than the "estimatedUSD".
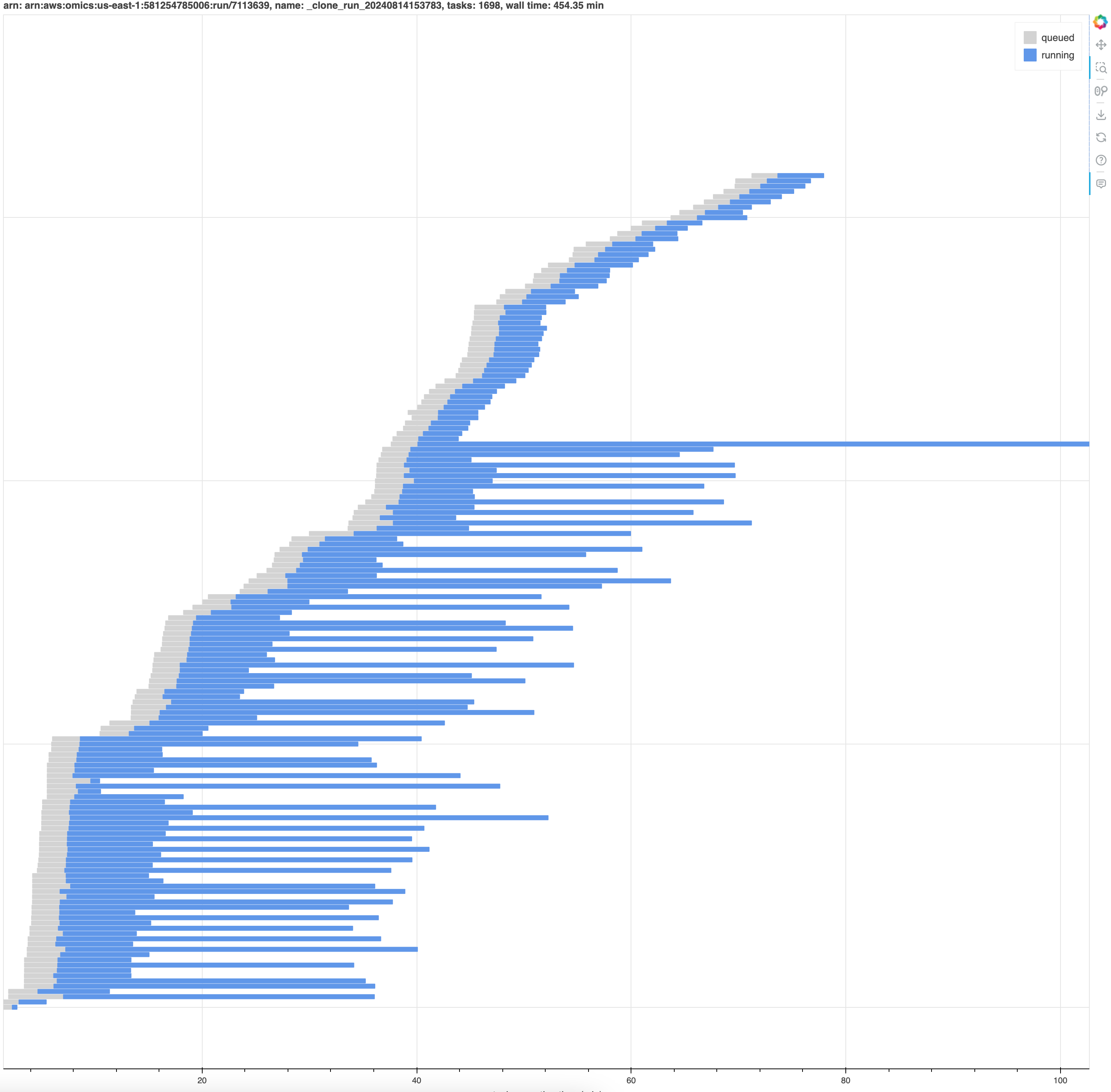
The RunAnalyzer tool can produce an interative timeline plot of a workflow. The plots allow you to visualize how individual tasks ran over the course of the run.
python -m omics.cli.run_analyzer -P plots/ 7113639python -m omics.cli.run_analyzer 1234567 -s -o run-1234567.jsonthis returns something like:
omics-run-analyzer: wrote run-1234567.json
See CONTRIBUTING for more information.
This project is licensed under the Apache-2.0 License.