MiranDa: Mimicking the Learning Process of Human Doctors to Achieve Causal Inference for Medication Recommendation
This is the first study to directly utilize causality in guiding parameter updates, providing highly accurate counterfactual results along with medication recommendations.
For commercial inquiries, please contact us at nagatomi@med.tohoku.ac.jp
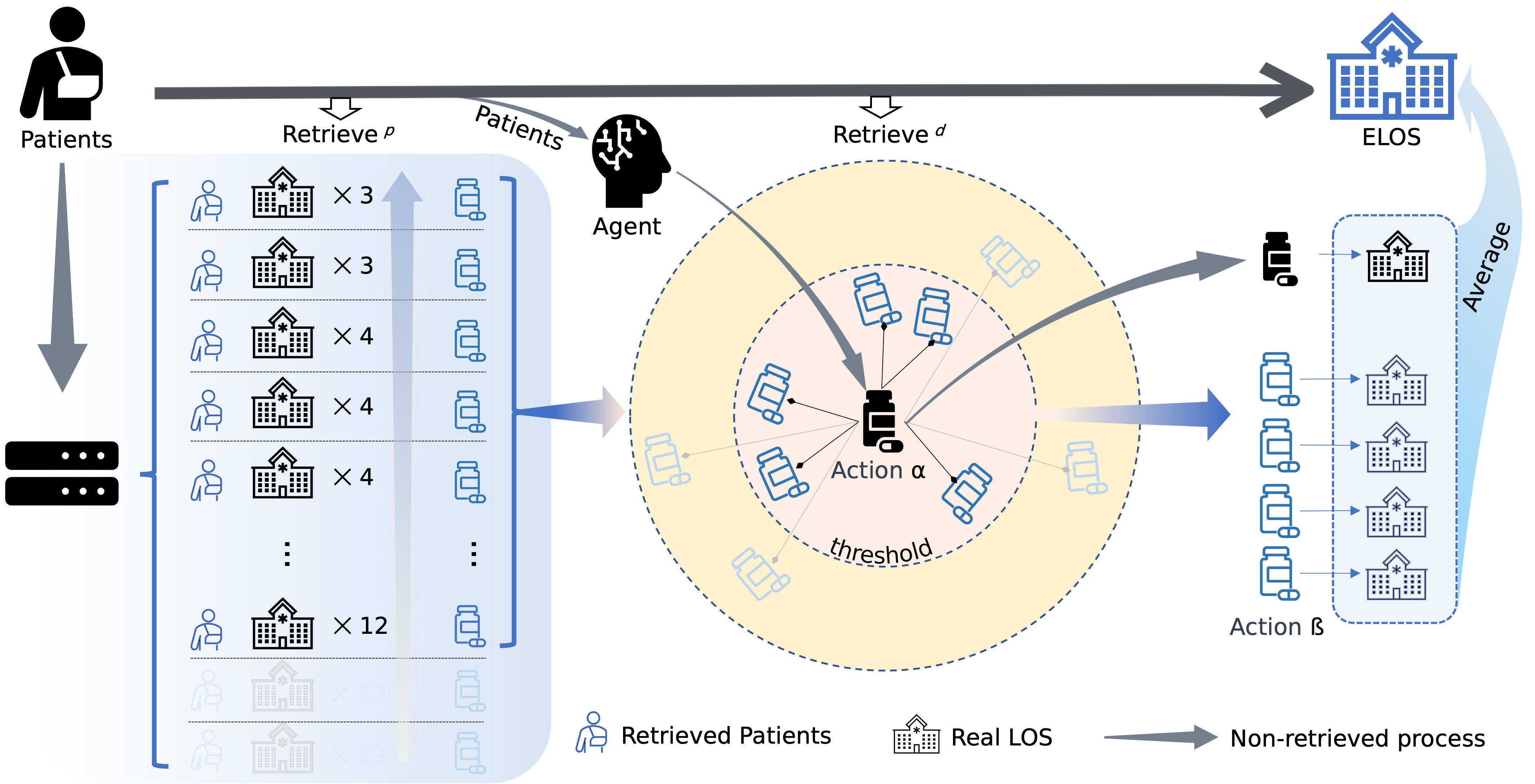
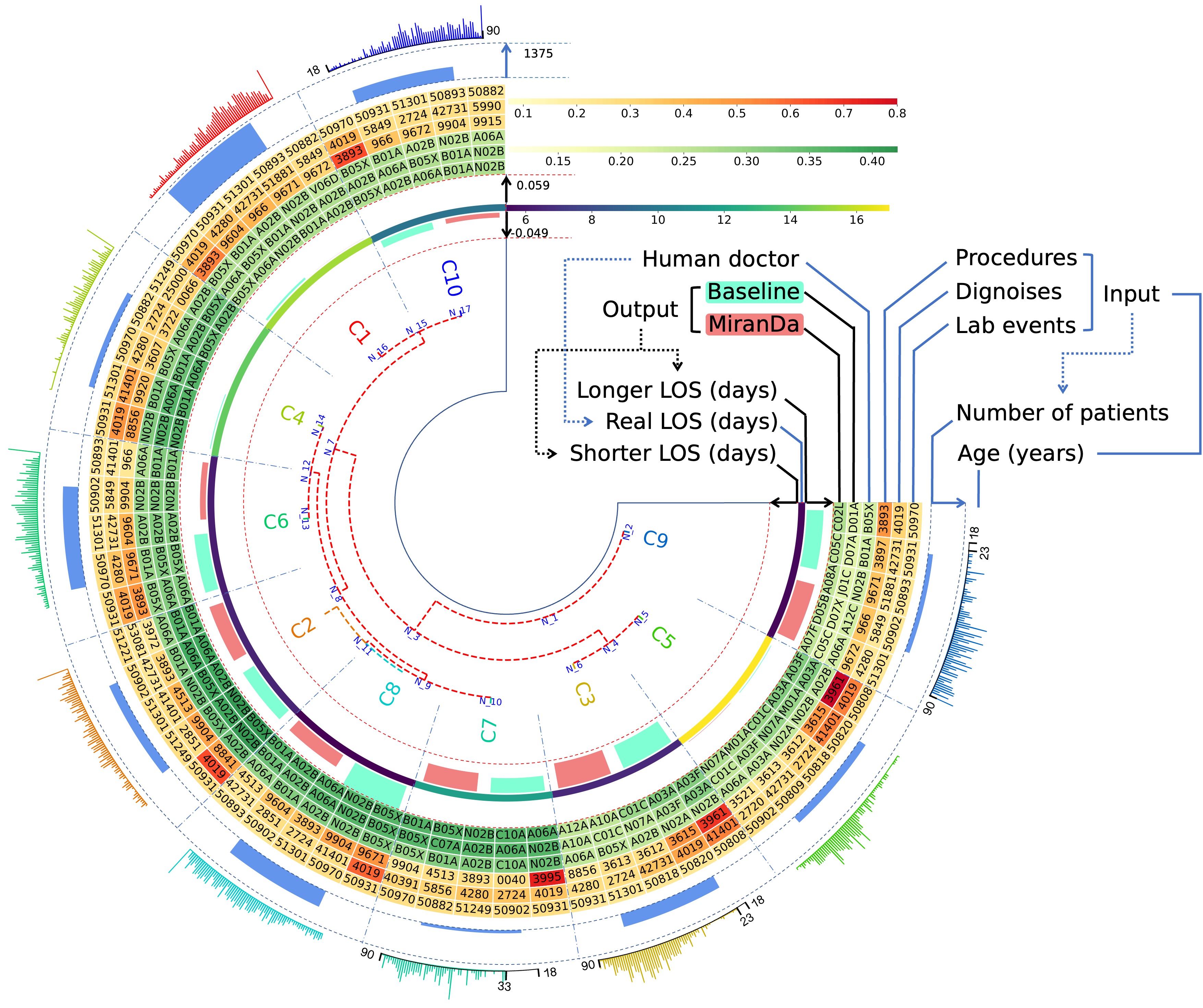 Starting from the center and moving outward, the first layer illustrates a Hierarchical Clustering Dendrogram with label colors indicating categories within the hyperbolic space and Poincaré model. The next three layers delineate the number of days: fewer than the patient's Length of Stay (LOS), equal to the LOS, and exceeding the LOS, respectively. The subsequent heatmap layer comprises six data sectors: MiranDa, baseline, human doctor, procedure, diagnosis, and lab events. The penultimate layer visualizes the number of individuals within the reconfigured group, and finally, the outermost layer presents the corresponding age distribution.
Starting from the center and moving outward, the first layer illustrates a Hierarchical Clustering Dendrogram with label colors indicating categories within the hyperbolic space and Poincaré model. The next three layers delineate the number of days: fewer than the patient's Length of Stay (LOS), equal to the LOS, and exceeding the LOS, respectively. The subsequent heatmap layer comprises six data sectors: MiranDa, baseline, human doctor, procedure, diagnosis, and lab events. The penultimate layer visualizes the number of individuals within the reconfigured group, and finally, the outermost layer presents the corresponding age distribution.
All input data have been accessed through credential verification from public sources, including MIMIC III database and MIMIC IV database.
we utilized a range of random seed values from 0 to 29, facilitating 30 distinct dataset splits into training, validation, and test sets for MIMIC III database, and 0 for MIMIC IV database. The models underwent a training period of 50 epochs, with an early stopping mechanism initiated from the fifth epoch, set to withstand a tolerance of three epochs. Decay factor λ, threshold δ, Confidence γ, Blend factor ϵ was 0.9, 0.2, 0.5, 0.2. The learning rate is 1 × 10−3. A batch size of 512 was uniformly adhered to across the experiment
python MiranDa.pyYou can adjust different hyperparameters in the cfg.py file.
| Hypermeters | Description |
|---|---|
dataset_list |
Train models for different datasets |
train_all_models |
Train different agents |
save_memory |
Delete useless features |
read_data |
Skip the preprocessing |
train_the_last |
I guess it's important, but I keep forgetting. If anyone is interested, please let me know. |
The development of this code was largely facilitated by pyHealth. Thanks!
I've been a huge fan of the Biohazard games, so I named this model after one of the characters from RE8.
If you have any questions, please let me know. E-mail: wang@med.tohoku.ac.jp