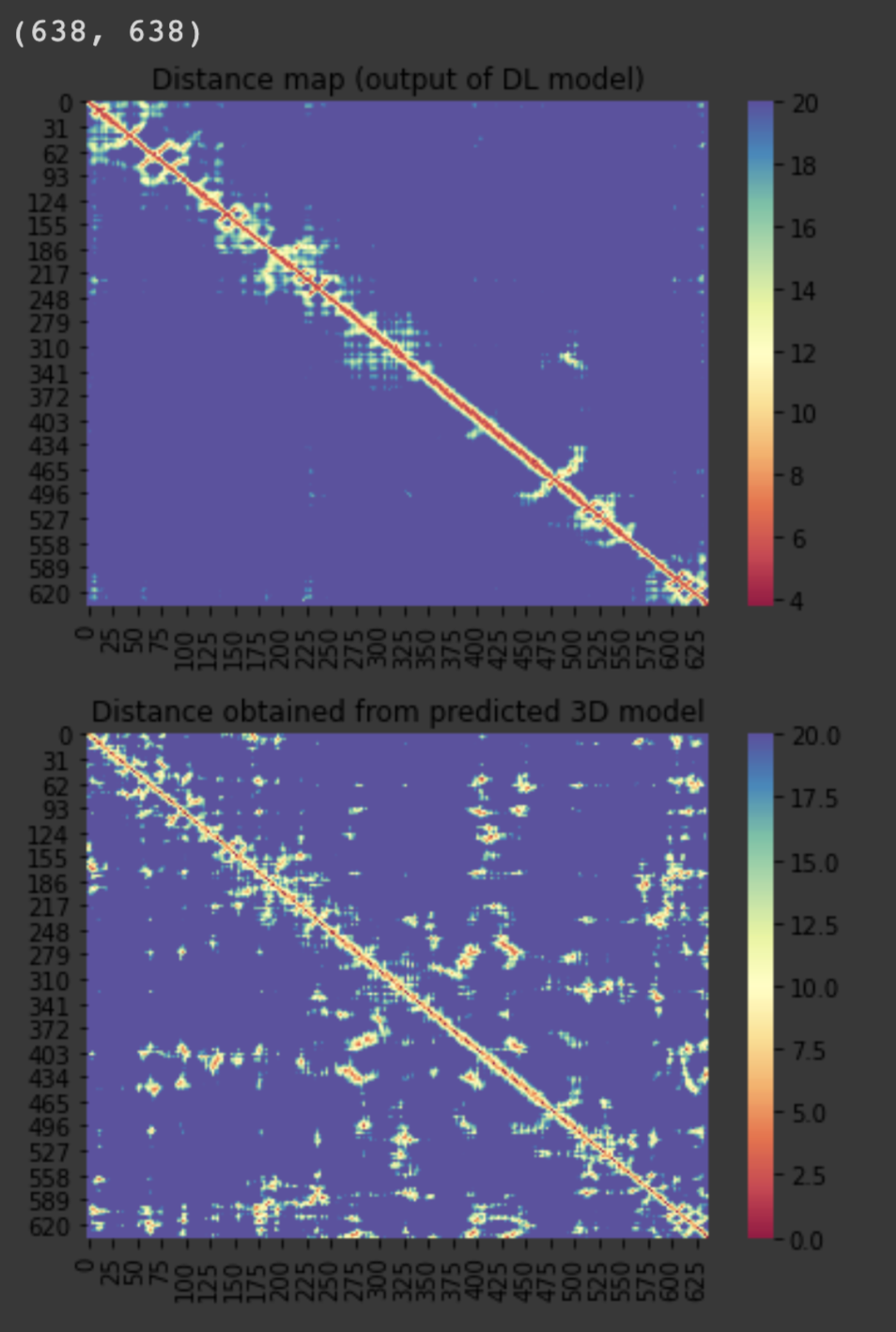
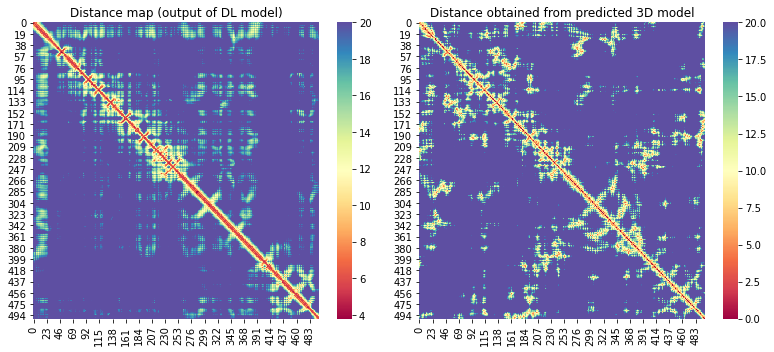
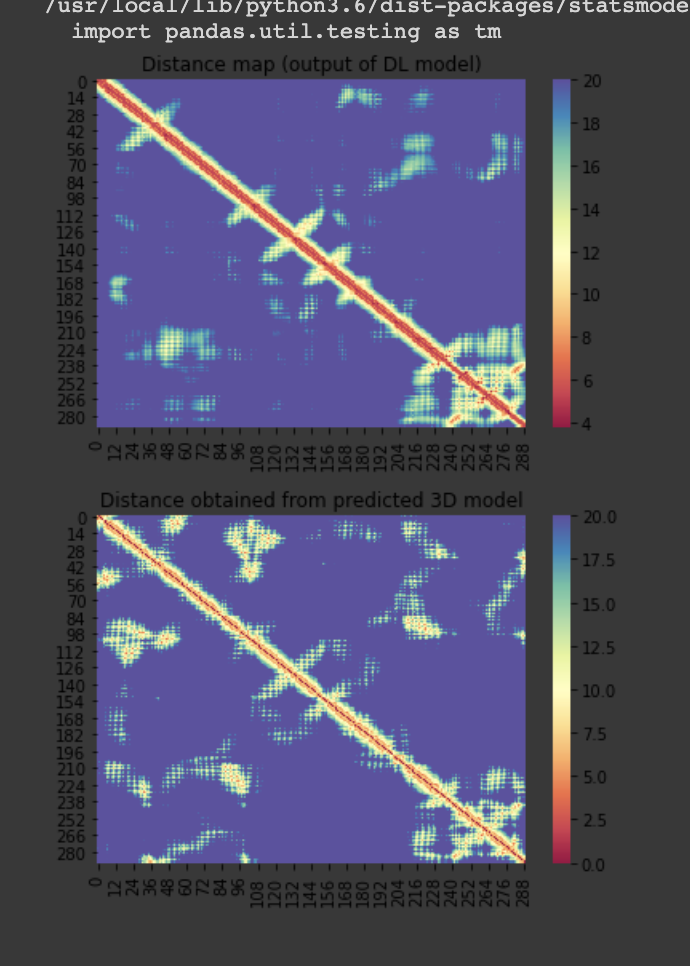
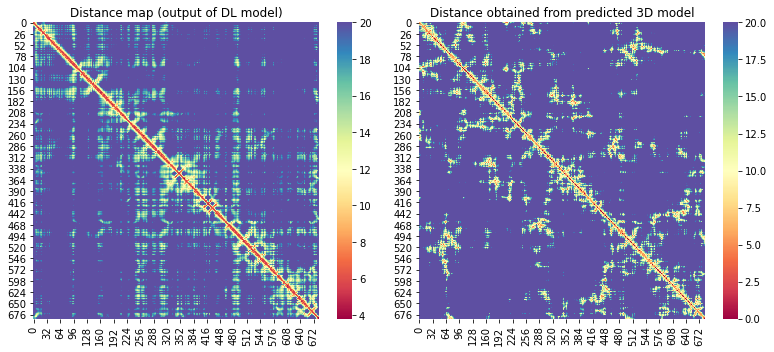
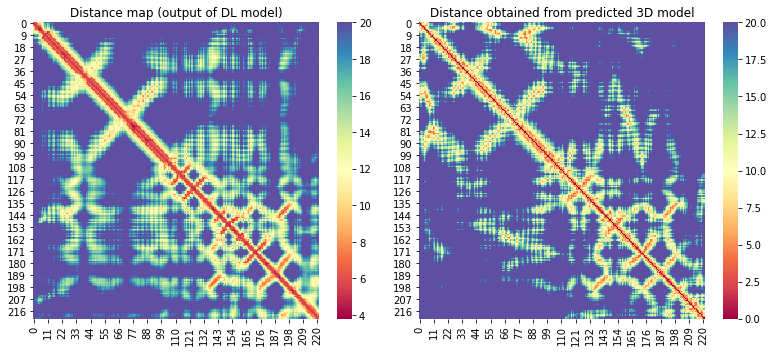
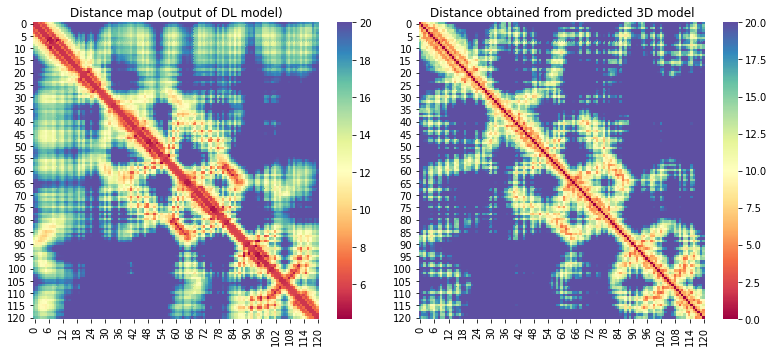
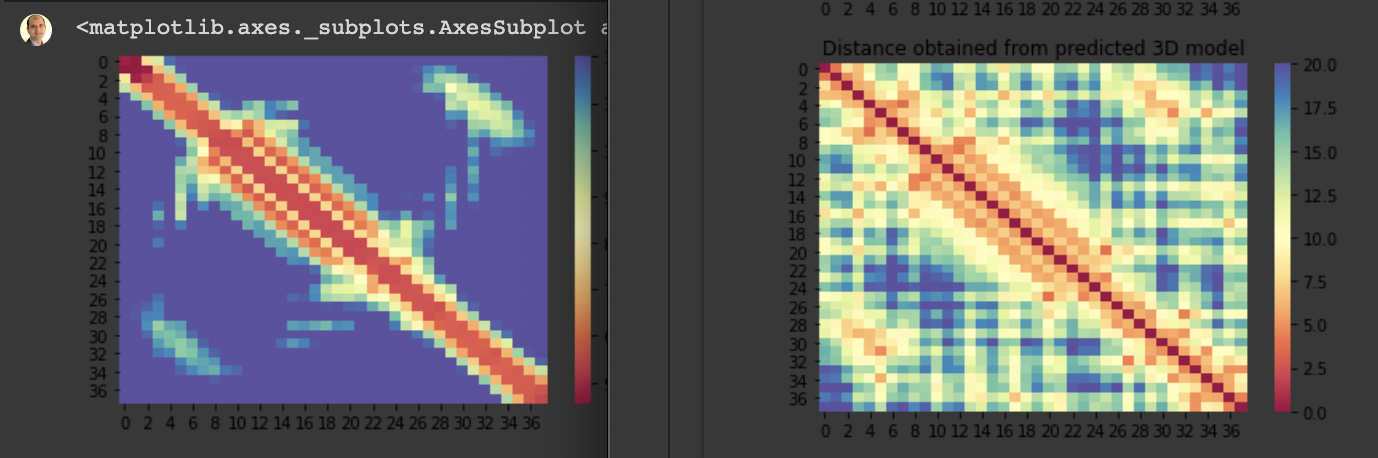
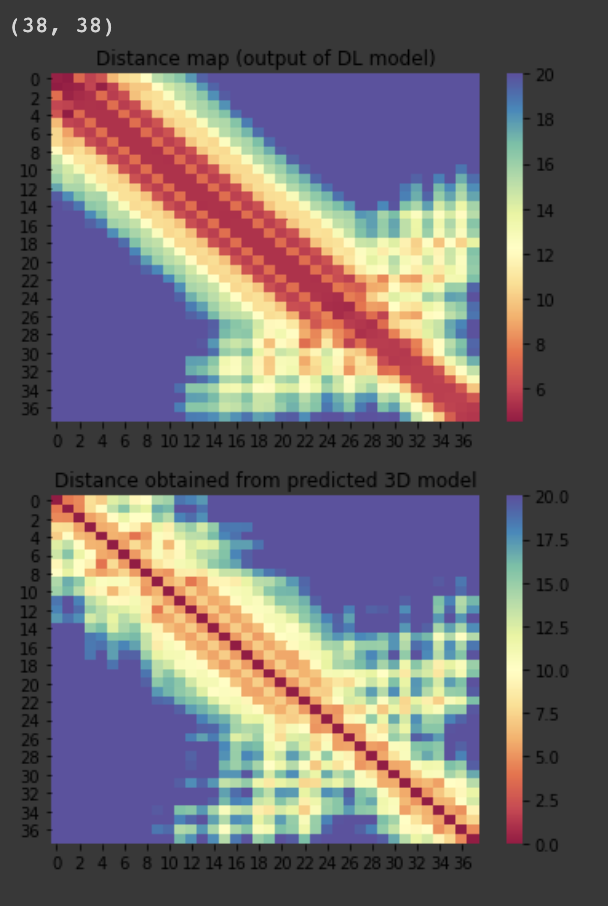
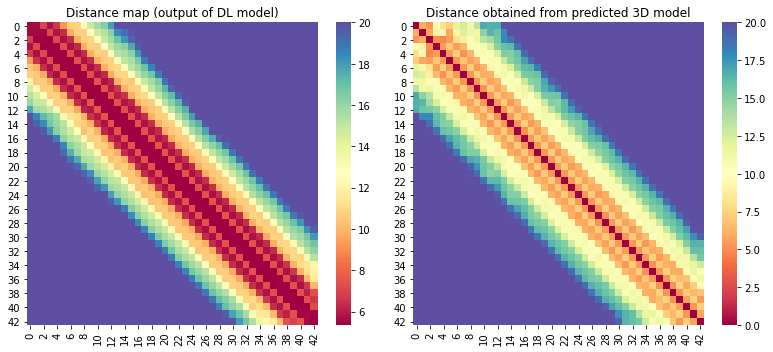
For the 10 COVID-19 related proteins released by CASP Commons
- Our overall method is demonstrated here
- Full details are yet to be published
| Protein (target) | L | Aln. size (e-value = 0.001) | Aln. size (e-value = 0.1) | # of models |
|---|---|---|---|---|
| C1901 | 638 | 249 | - | 25 |
| C1902 | 500 | 499 | - | 17 |
| C1903 | 290 | 357 | - | 150 |
| C1904 | 686 | 166 | - | 12 |
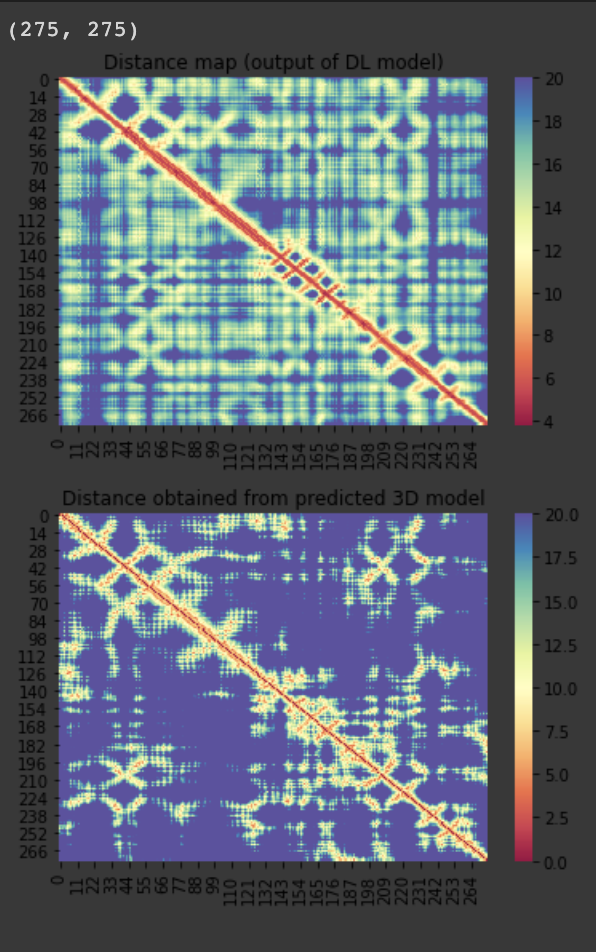
| C1905 | 275 | 26 | - | 200 |
| C1906 | 222 | 160 | - | 100 |
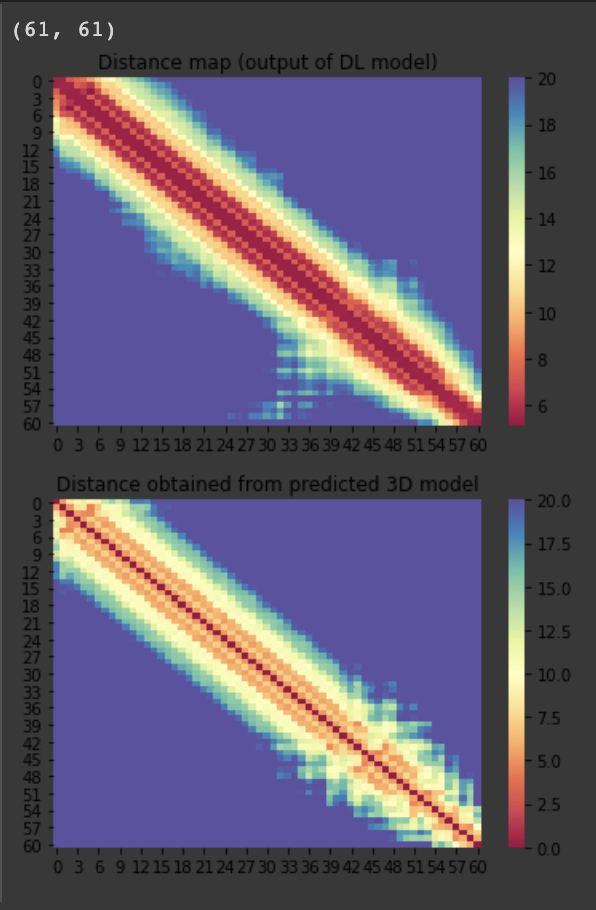
| C1907 | 61 | 12 | - | 350 |
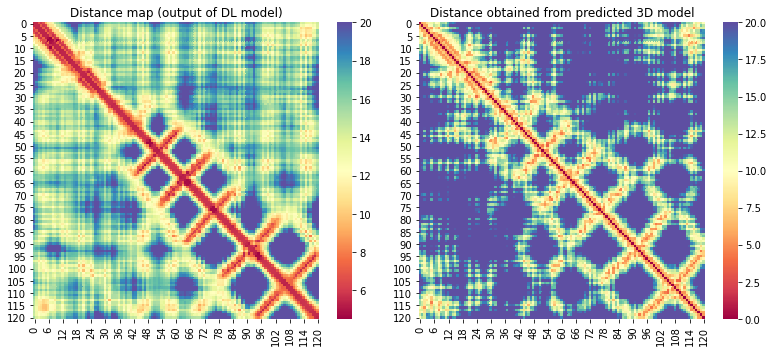
| C1908 | 121 | 12 | 1919 | 420 |
| C1909 | 38 | 1 | 1 | 600 |
| C1910 | 43 | 4 | 2141 | 410 |
- Badri Adhikari (adhikarib@umsl.edu)
- Jacob Barger (jsbp67@mail.umsl.edu)
- Bikash Shrestha (bsmmy@mail.umsl.edu)
Badri Adhikari
adhikarib@umsl.edu
University of Missouri-St. Louis