This repository includes code on the development of a 3D Unet on MRI scan images containing 4 modalities (T2 Flair, T1w, T1Gd and T2w). The output of the model includes 3 segmentations (Whole tumor, Tumor core and Enhancing tumor)
The dataset is a subset of the Brats 16/17 challenge and is available at http://medicaldecathlon.com/. In total the dataset contains 484 MRI scans with all 4 modalities segmented for edema (label 1) , non enhancing tumor (label 2) and enhancing tumor core (label 3).
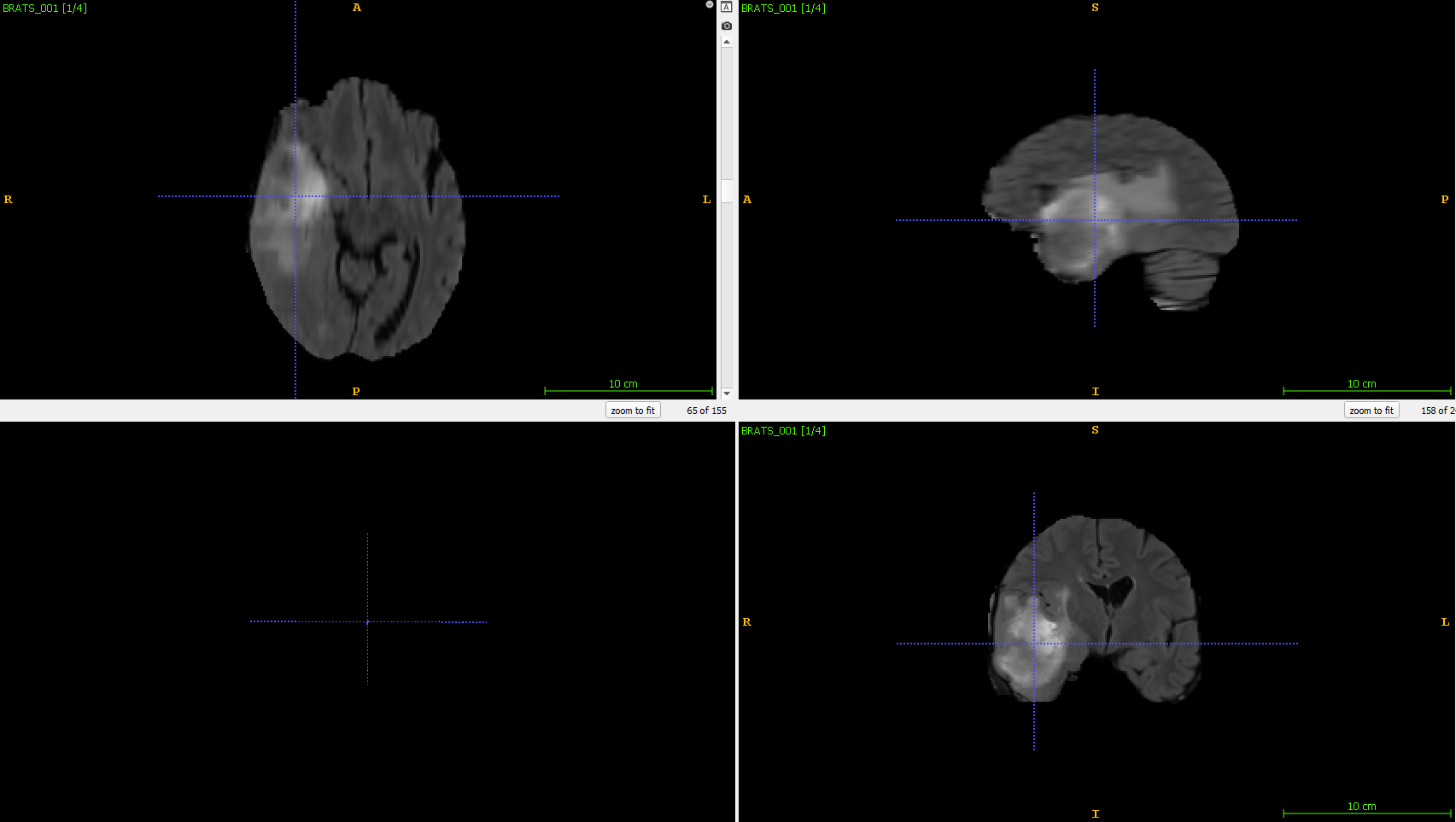
An illustration of the T2 Flair modality can be seen below:
The folder structure of this repository is shown below:
The 'data' folder contains the original dataset with seperate sub-folders for training images (imagesTr), training labels (labelsTr) and test images (imagesTs). The images and segmentation labels are in .nii.gz format.
The 'multi_modal_3D_brain_tumor_segmentation.ipynb' notebook is used to pre-process, split the training data into train and validation sets and to train the 3D Unet model. The project monai (https://monai.io/) was used to construct the 3D Unet with minimal effort. All code is written in python with the Pytorch deep learning framework.
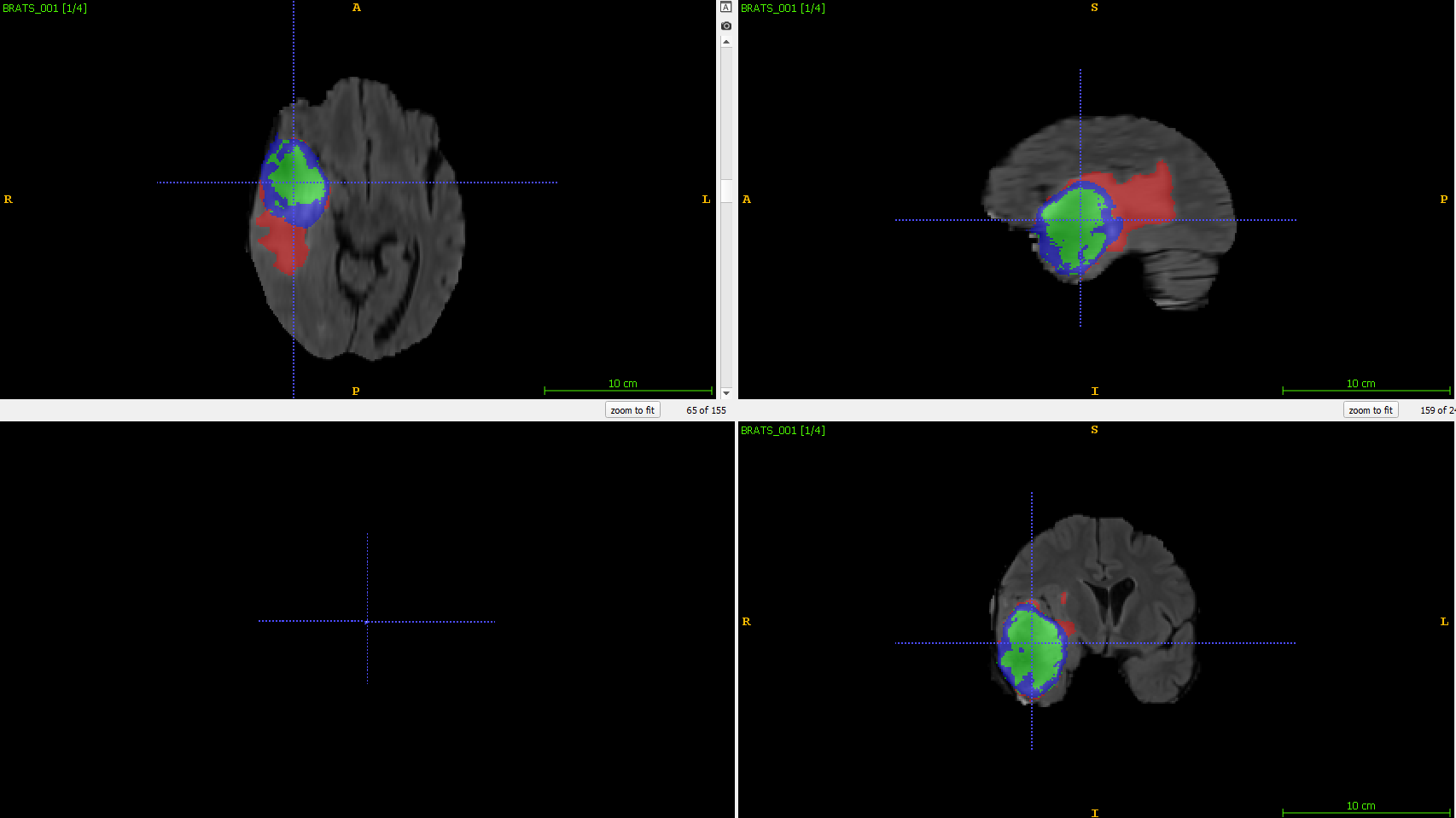
An example of the ground truth (left) and the prediction (right) is shown below:
| Original image | 3D Unet segmented image |
|---|---|
 |
 |
@software{nic_ma_2021_4891800, author = {Nic Ma and Wenqi Li and Richard Brown and Yiheng Wang and Behrooz and Benjamin Gorman and Hans Johnson and Isaac Yang and Eric Kerfoot and charliebudd and Yiwen Li and Mohammad Adil and Yuan-Ting Hsieh (謝沅廷) and Arpit Aggarwal and Cameron Trentz and adam aji and masadcv and Mark Graham and Ben Murray and Gagan Daroach and Petru-Daniel Tudosiu and myron and Matt McCormick and Ambros and Balamurali and Christian Baker and Jan Sellner and Lucas Fidon and cgrain}, title = {Project-MONAI/MONAI: 0.5.3}, month = jun, year = 2021, publisher = {Zenodo}, version = {0.5.3}, doi = {10.5281/zenodo.4891800}, url = {https://doi.org/10.5281/zenodo.4891800} }
@misc{simpson2019large, title={A large annotated medical image dataset for the development and evaluation of segmentation algorithms}, author={Amber L. Simpson and Michela Antonelli and Spyridon Bakas and Michel Bilello and Keyvan Farahani and Bram van Ginneken and Annette Kopp-Schneider and Bennett A. Landman and Geert Litjens and Bjoern Menze and Olaf Ronneberger and Ronald M. Summers and Patrick Bilic and Patrick F. Christ and Richard K. G. Do and Marc Gollub and Jennifer Golia-Pernicka and Stephan H. Heckers and William R. Jarnagin and Maureen K. McHugo and Sandy Napel and Eugene Vorontsov and Lena Maier-Hein and M. Jorge Cardoso}, year={2019}, eprint={1902.09063}, archivePrefix={arXiv}, primaryClass={cs.CV} }