RegGNN, a graph neural network architecture for many-to-one regression tasks with application to functional brain connectomes for IQ score prediction, developed in Python by Mehmet Arif Demirtaş (demirtasm18@itu.edu.tr). This work has been published in Brain Imaging and Behavior.
This repository contains the implementation for the proposed RegGNN model and learning-based sample selection, and the codes for the comparison models from the following paper:
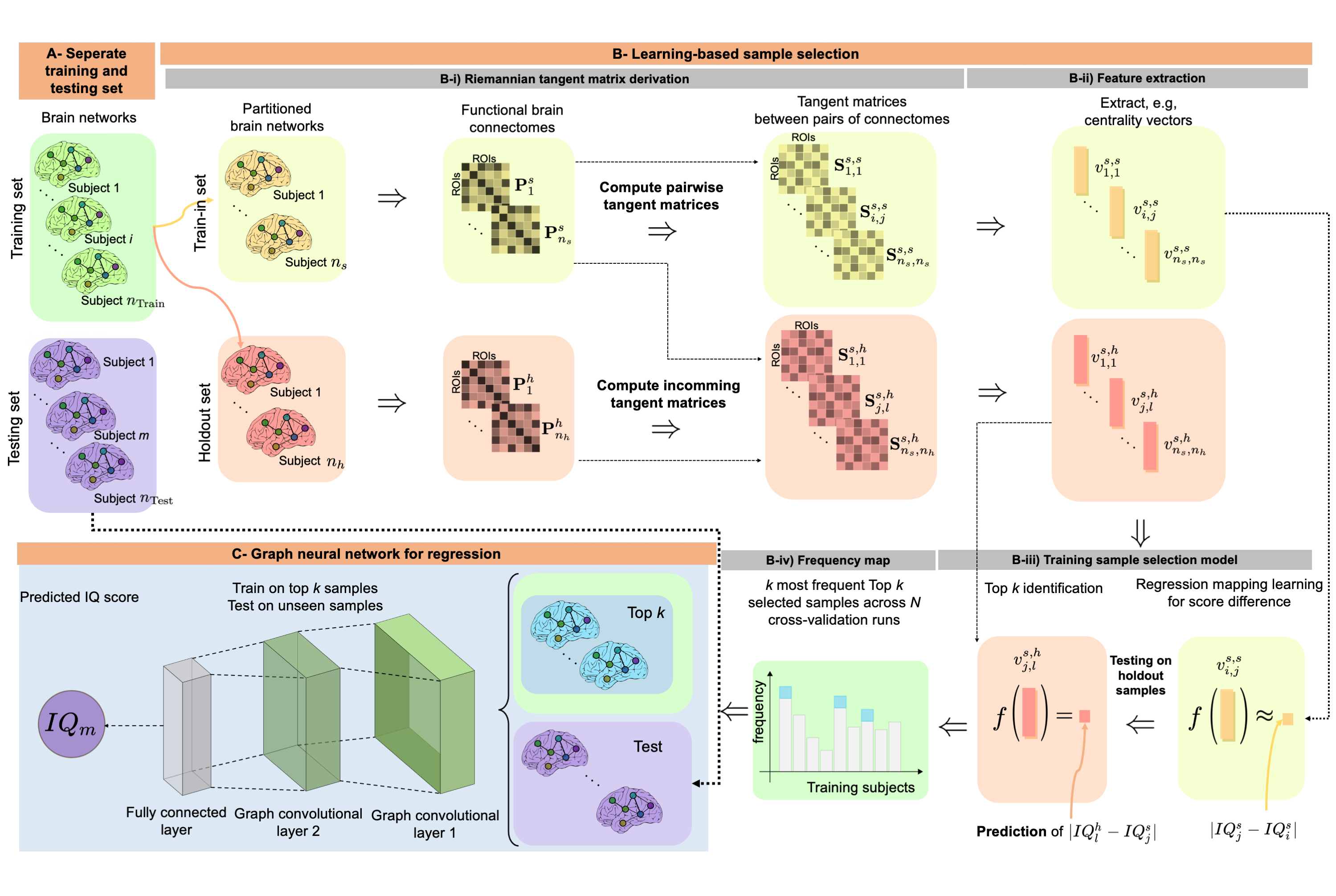
Predicting cognitive scores with graph neural networks through sample selection learning
Martin Hanik†1, Mehmet Arif Demirtaş†2, Mohammed Amine Gharsallaoui2, 3, Islem Rekik2
1Zuse Institute Berlin, Berlin, Germany
2BASIRA Lab, Faculty of Computer and Informatics, Istanbul Technical University, Istanbul, Turkey
3Ecole Polytechnique de Tunisie (EPT), Tunis, Tunisia
†Equal ContributionAbstract: Analyzing the relation between intelligence and neural activity is of the utmost importance in understanding the working principles of the human brain in health and disease. In existing literature, functional brain connectomes have been used successfully to predict cognitive measures such as intelligence quotient (IQ) scores in both healthy and disordered cohorts using machine learning models. However, existing methods resort to flattening the brain connectome (i.e., graph) through vectorization which overlooks its topological properties. To address this limitation and inspired from the emerging graph neural networks (GNNs), we design a novel regression GNN model (namely RegGNN) for predicting IQ scores from brain connectivity. On top of that, we introduce a novel, fully modular sample selection method to select the best samples to learn from for our target prediction task. However, since such deep learning architectures are computationally expensive to train, we further propose a learning-based sample selection method that learns how to choose the training samples with the highest expected predictive power on unseen samples. For this, we capitalize on the fact that connectomes (i.e., their adjacency matrices) lie in the symmetric positive definite (SPD) matrix cone. Our results on full-scale and verbal IQ prediction outperforms comparison methods in autism spectrum disorder cohorts and achieves a competitive performance for neurotypical subjects using 3-fold cross-validation. Furthermore, we show that our sample selection approach generalizes to other learning-based methods, which shows its usefulness beyond our GNN architecture.
- Code for RegGNN and sample selection methods is written by Mehmet Arif Demirtaş.
- Code for CPM model is written by esfinn, our functions are taken from this notebook.
- Code for PNA model is written by lukecavabarrett, available in this repository.
- Code for SPD, geometric processing tool, is developed within the geometric data analysis and processing research group at Zuse Institute Berlin and is available at Morphomatics.
Code for RegGNN was prepared using Python 3.7.10. The major dependencies are listed below, and the full list can be found in requirements.txt:
- torch >= 1.7.0
- torch_scatter >= 2.0.6
- torch_sparse >= 0.6.9
- torch_cluster >= 1.5.9
- torch_spline_conv >= 1.2.1
- torch_geometric >= 1.6.3
- scikit-learn >= 0.24.1
- pymanopt >= 0.2.5
- numpy >= 1.19.5
- pandas >= 1.1.5
- scipy >= 1.5.4
Most packages can be installed using pip install -r requirements.txt command, whereas torch_scatter, torch_sparse, torch_cluster and torch_spline_conv can be installed from using the following commands, as they are not available on PyPI:
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-1.7.0+cu102.html
pip install torch-sparse -f https://pytorch-geometric.com/whl/torch-1.7.0+cu102.html
pip install torch-cluster -f https://pytorch-geometric.com/whl/torch-1.7.0+cu102.html
pip install torch-spline-conv -f https://pytorch-geometric.com/whl/torch-1.7.0+cu102.html
Detailed information on torch_geometric installation can be found on their website.
You can directly download the functional connectomes at: http://fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html
demo.py file can be used to either prepare real data for usage in RegGNN by extracting topological features, or it can be used to create simulated data and topological features using the parameters specified in config.py file.
To simulate functional brain connectome data and create topological features using eigenvector centrality measure, use the following command:
python demo.py --mode data --data-source simulated --measure eigenTo extract topological features using node degree centrality measure from existing data, use the following command:
python demo.py --mode data --data-source saved --measure nodeAll valid values for measures are:
abs: Absolute values between connectomesgeo: Scalar Riemannian distance valuetan: Featurized tangent matrixeigen: Eigenvector centralitynode: Node degree centralityclose: Closeness centralityconcat_orig: Concatenated centralities (not scaled)concat_scale: Concatenated centralities (min-max scaled)
demo.py file can also be used to apply cross-validation on prepared data using the preprocess commands above, using the parameters specified in config.py file. In addition to our proposed RegGNN model, other comparison models used in the paper, CPM and PNA are also supported.
To use RegGNN model and sample selection to make inferences on data:
python demo.py --mode infer --model RegGNNDifferent options for models and data can be specified using config.py file. The options used throughout the program are as follows:
System options:
DATA_FOLDER: path to the folder data will be written to and read fromRESULT_FOLDER: path to the folder data will be written to and read from
Simulated data options:
CONNECTOME_MEAN: mean of the distribution from which connectomes will be sampledCONNECTOME_STD: std of the distribution from which connectomes will be sampledSCORE_MEAN: mean of the distribution from which scores will be sampledSCORE_STD: std of the distribution from which scores will be sampledN_SUBJECTS: number of subjects in the simulated dataROI: number of regions of interest in brain graphSPD: whether or not to make generated matrices symmetric positive definite
Evaluation options:
K_FOLDS: number of cross validation folds
RegGNN options:
NUM_EPOCH: number of epochs the process will be run forLR: learning rateWD: weight decayDROPOUT: dropout rate
PNA options:
NUM_EPOCH: number of epochs the process will be run forLR: learning rateWD: weight decayDROPOUT: dropout rateSCALERS: scalers used by PNAAGGRS: aggregators used by PNA
Sample selection options:
SAMPLE_SELECTION: whether or not to apply sample selectionK_LIST: list of k values for sample selectionN_SELECT_SPLITS: number of folds for the nested sample selection cross validation
Randomization options:
DATA_SEED: random seed for data creationMODEL_SEED: random seed for modelsSHUFFLE: whether to shuffle or not
@article{hanik2022predicting,
title={Predicting cognitive scores with graph neural networks through sample selection learning},
volume={16},
ISSN={1931-7557, 1931-7565},
url={https://link.springer.com/10.1007/s11682-021-00585-7},
DOI={10.1007/s11682-021-00585-7},
number={3},
journal={Brain Imaging and Behavior},
author={Hanik, Martin and Demirtaş, Mehmet Arif and Gharsallaoui, Mohammed Amine and Rekik, Islem},
year={2022},
month={Jun},
pages={1123–1138},
language={en}
}Link: https://arxiv.org/abs/2106.09408
Our code is released under MIT License (see LICENSE file for details).