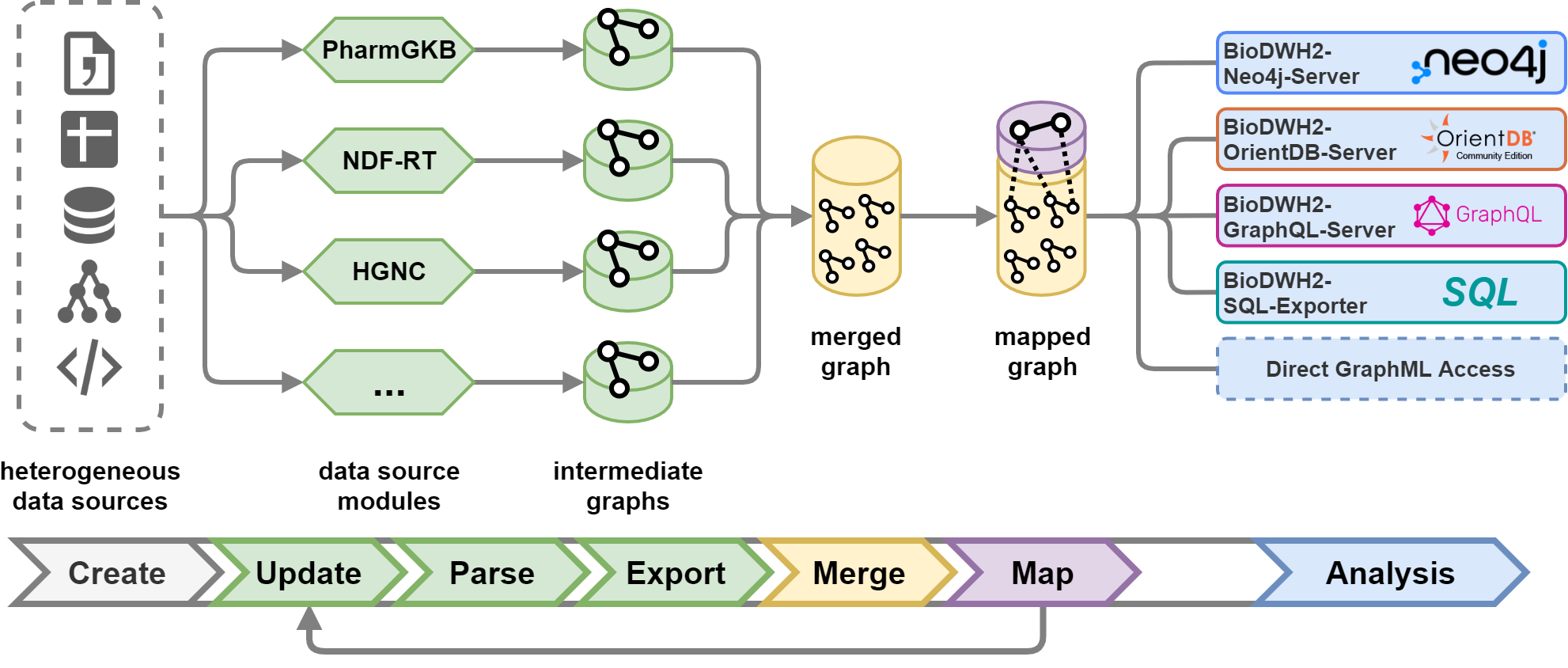
BioDWH2 is an easy-to-use, automated, graph-based data warehouse and mapping tool for bioinformatics and medical informatics. Whether you need a data warehouse for running research analyses or information systems, BioDWH2 can support you in setting things up.
Multiple data sources are readily available with many more under development or planned. A list of officially supported data sources is provided here.
Missing a data source important to you? Feel free to open a discussion here! Want to develop your own data source module? In that case consult the development documentation.
The latest release version of BioDWH2 can be downloaded here.
A list of changes for each version can be found in the Changelog.
For using generated data warehouses inside Neo4j please see the separate BioDWH2-Neo4j-Server repository.
BioDWH2 is developed to be usable out of the box without any prerequisites except the Java Runtime Environment version 8. The JRE 8 is available here.
For detailed information on how to use BioDWH2 please see the separate documentation.
If you either want to help in the development of BioDWH2 directly, or write a new data source module please consult the development documentation.
If you make use of BioDWH2 or it's companion tools as part or your research cite the BioDWH2 manuscript in any resulting publications.
Friedrichs, M. (2021). BioDWH2: an automated graph-based data warehouse and mapping tool. Journal of Integrative Bioinformatics. https://doi.org/10.1515/jib-2020-0033