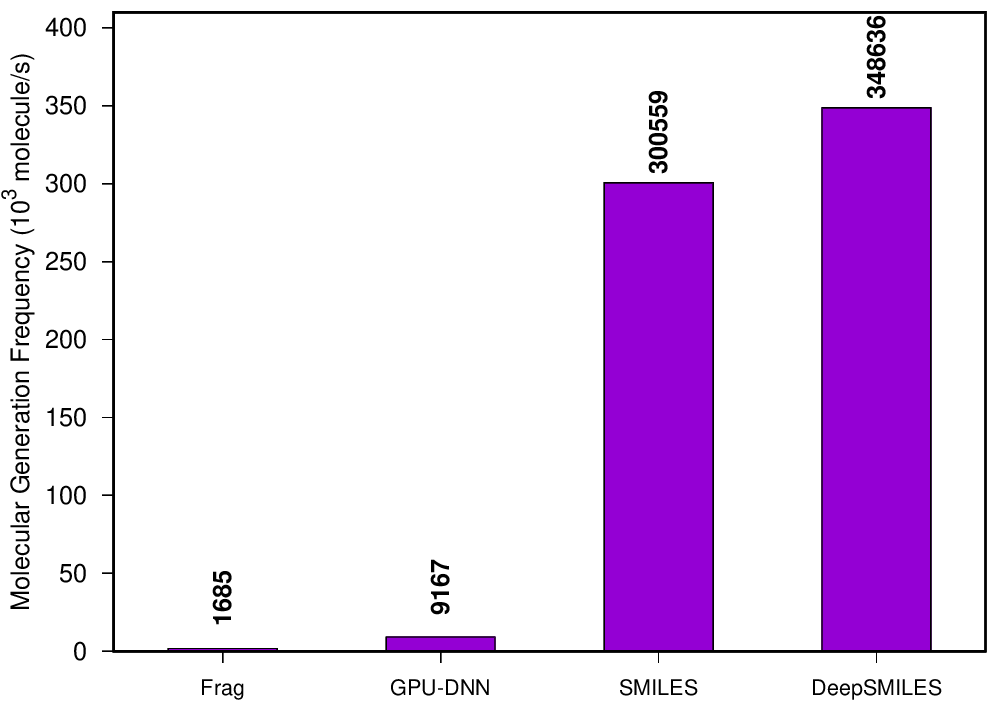
Reference implementation for the article "Molecular Generation by Fast Assembly of (Deep)SMILES Fragments". Generate molecules fast from a molecular training set while also doing training-set distribution matching.
On Linux, you need python3-rdkit to be installed.
For OCaml programmers, you can clone this repository then type 'make && make install'. Note that you need to have opam installed and configured.
For end-users:
sudo apt install opam
opam init
eval `opam config env`
opam install fasmifraFor programmer end-users, or if the opam package is not ready yet:
sudo apt install opam
opam init
eval `opam config env`
opam pin add fasmifra https://github.com/UnixJunkie/FASMIFRA.gitWe are currently working on an automated self-installer; stay tuned.
Those molecules are your "molecular training set".
fasmifra_fragment.py -i my_molecules.smi -o my_molecules_frags.smiIf you fragment rather small molecules, you might want to use the -w option and pass a smaller recommended fragment weight than the default (150 Da).
usage: fasmifra_fragment.py [-h] [-i input.smi] [-o output.smi] [--seed SEED]
[-n NB_PASSES] [-w FRAG_WEIGHT]
fragment molecules (tag cleaved bonds)
optional arguments:
-h, --help show this help message and exit
-i input.smi molecules input file
-o output.smi fragments output file
--seed SEED RNG seed
-n NB_PASSES number of fragmentation passes
-w FRAG_WEIGHT fragment weight (default=150Da)
fasmifra -n 100000 -i my_molecules_frags.smi -o my_molecules_gen.smiusage:
fasmifra
-n <int>: how many molecules to generate
-i <filename>: smiles fragments input file
-o <filenams>: output file
[--seed <int>]: RNG seed
[--deep-smiles]: input/output molecules in DeepSMILES no-rings format
| Benchmark | Random sampler | SMILES LSTM | Graph MCTS | AAE | ORGAN | VAE | FASMIFRA | Negative control |
|---|---|---|---|---|---|---|---|---|
| Validity | 1.000 | 0.959 | 1.000 | 0.822 | 0.379 | 0.870 | 1.000 | 1.000 |
| Uniqueness | 0.997 | 1.000 | 1.000 | 1.000 | 0.841 | 0.999 | 0.994 | 0.959 |
| Novelty | 0.000 | 0.912 | 0.994 | 0.998 | 0.687 | 0.974 | 0.702 | 0.947 |
| KL_divergence | 0.998 | 0.991 | 0.522 | 0.886 | 0.267 | 0.982 | 0.959 | 0.855 |
| FCD | 0.929 | 0.913 | 0.015 | 0.529 | 0.000 | 0.863 | 0.814 | 0.397 |
[1] Berenger, F., Tsuda, K. Molecular generation by Fast Assembly of (Deep)SMILES fragments. J Cheminform 13, 88 (2021). https://doi.org/10.1186/s13321-021-00566-4
[2] O'Boyle, N., & Dalke, A. (2018). "DeepSMILES: an adaptation of SMILES for use in machine-learning of chemical structures". chemrxiv.org
[3] Weininger, D. (1988). SMILES, a chemical language and information system. "1. Introduction to methodology and encoding rules". Journal of chemical information and computer sciences, 28(1), 31-36.