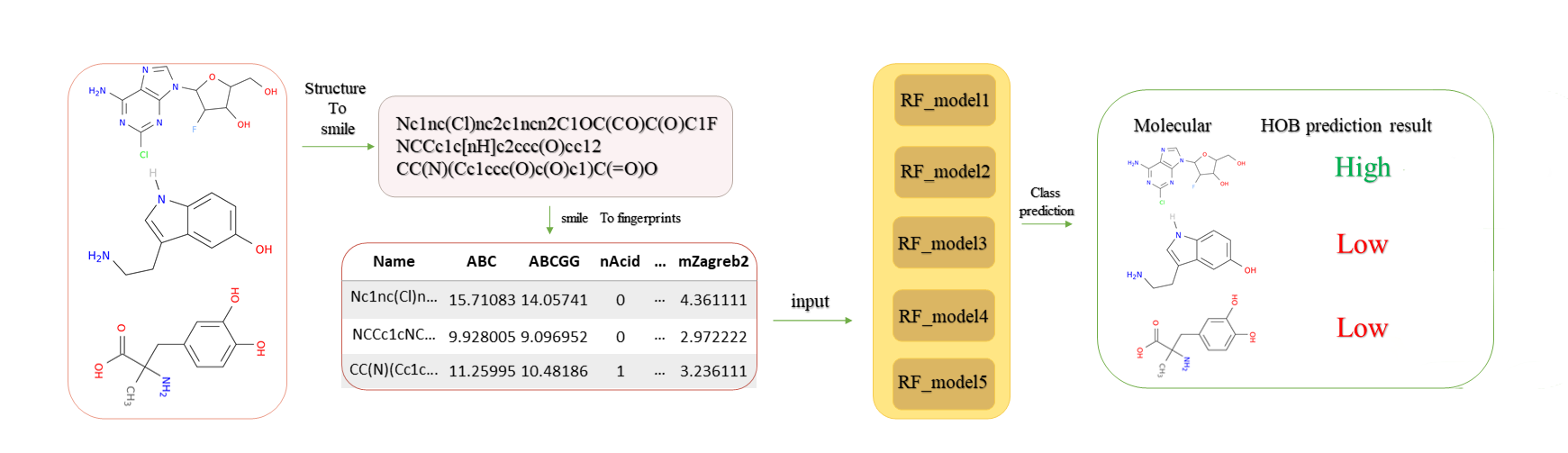
Predictions can also be made directly on our online server at http://www.icdrug.com/ICDrug/A
Conda environment
$ conda HobPre create -f Hob_env.yml
Python 3.6
-
Mordred==1.2.0
-
scikit-learn==0.23.2
-
pandas==1.1.1
-
numpy==1.19.2
-
matplotlib==3.3.4
Usage
$ conda activate HobPre
$ python HOB_predict.py your_model_path your_smiles.txt cutoff
example:
$ python HOB_predict.py model smiles.txt 20
Parameter meaning:
model_path:The folder where the model to be used for prediction is located
smiles.txt:input smiles file,one per each line
cutoff: 50. Use F=50% as the cut-off value. If F>50%, the predicted oral availability is qualitatively high.
20. Use F=20% as the cut-off value. If F>20%, the predicted oral availability is qualitatively high.
Reference
Min Wei, Xudong Zhang, Xiaolin Pan, Bo Wang, Changge Ji, Yifei Qi, and John Z.H. Zhang.HobPre: accurate prediction of human oral bioavailability for small molecules (submitted)
*The data used in this paper can be obtained from hob_data_set.xlsx
Model Parameters License
The HOB prediction models are made available for non-commercial use only, under the terms of the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. You can find details at: https://creativecommons.org/licenses/by-nc/4.0/legalcode