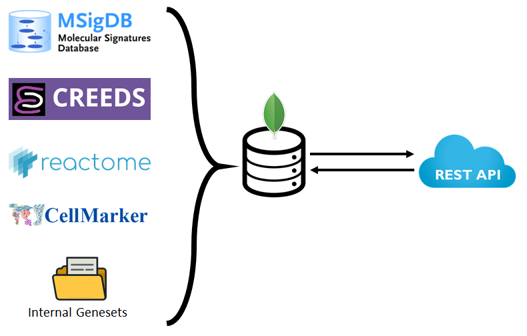
- Central repository for storing geneset references
- Introduce new GMTx file format for storing geneset-related data
- Provide programmatic access to the database with a RESTful API
- Albert Kang (swk30@cam.ac.uk)
- Laura Badi (badilla@gmx.ch)
-
Like GMT files:
- It is a tab-delimited text file
- The trailing tail columns represent the membership genes
-
Unlike GMT files:
- GMTx files allow metadata on top of geneset names and geneset descriptions
- GMTx files allow coefficients attached to a membership gene to be stored
-
See the explanation of the GMT file format here: https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats#GMT:_Gene_Matrix_Transposed_file_format_.28.2A.gmt.29
-
GMTx file format on top of the GMT file format is as follows:
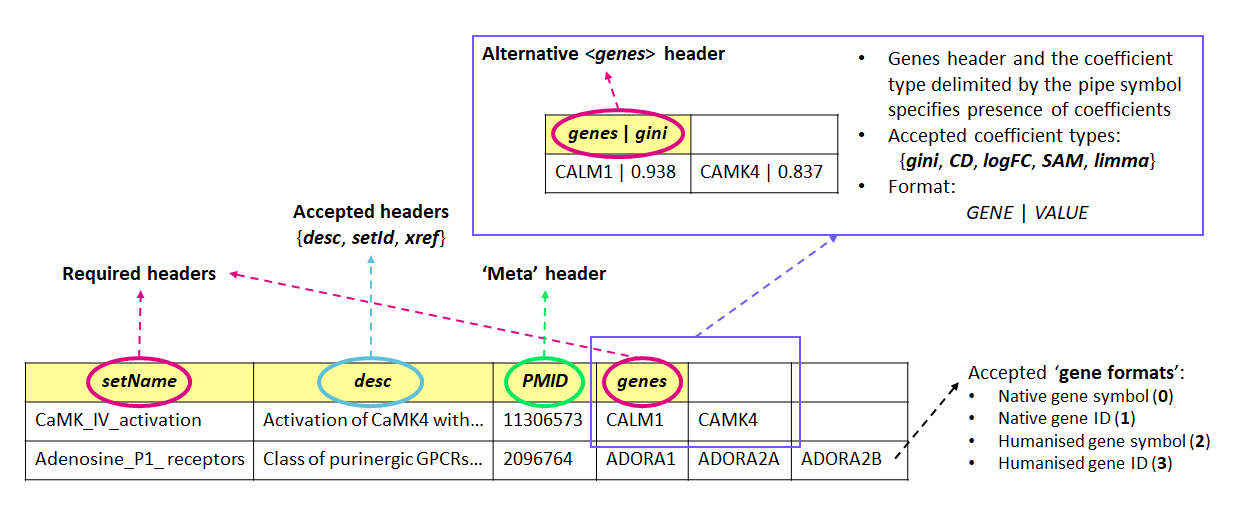
- GMTx files require row 1 to be contain column headers:
- ...accepted headers are:
setName,genes,xref,setId,desc - ... any other header names will be considered as a 'meta-tag'
- ...accepted headers are:
- GMTx files MUST include the following headers:
setNameandgenes- ...
setNameSHOULD be the first header column - ...
genesMUST be the last header column
- ...
- Membership gene can be written in the following formats:
- ...
GENE(as in regular GMT files) - ...
GENE | VALUE(new to GMTx files)
- ...
- GMTx files require row 1 to be contain column headers:
Instructions for converting to the GMTx file format can be found here: ./GeMS/src/x_to_gmtx_converter
pip install gunicorn
pip install pymongo
pip install flask
pip install flask_restful
pip install xmltodict
The main upload logic is here: \GeMS\src\api\upload.py
The CLI arguments are as follows:
| Args | Name | Required | Examples |
|---|---|---|---|
--fl |
File location | O | |
--gf |
Gene format | O | 0, 1, 2, 3 |
--so |
Source | O | Roche, MSigDB... |
--ti |
NCBI Taxonomy ID | O | 9606, 10090... |
--us |
User | O | Public, badil... |
--st |
Subtype | X | C7, BP... |
--do |
Domain | X | pathway, cell marker... |
[\GeMS\src\api\] python upload.py --fl ../../data/Reactome/ReactomePathways.gmtx --gf 0 --so Reactome --ti 9606 --us Public --do pathway
[\GeMS\] chmod +x upload.sh
[\GeMS\] sbatch -J bulkUpload -o bulkUpload.out -e bulkUpload.err --ntasks=1 --qos=normal --cpus-per-task=16 --wrap="./upload.sh"
You can query the genesets stored in GeMS using our REST-API service. A detailed explanation of the supported tools and services can be found here: ./GeMS/src/api
From version 1.4.0, ribiosGSEA supports reading, inserting, and removing genesets from GeMS. See the vignette 'working-with-GeMS' of the ribiosGSEA package.
.
├── ...
├── src
│ │
│ ├── api Flask REST-API and GMTx file loader
│ │ │
│ │ ├── db_utils.py Database configuration
│ │ │
│ │ ├── app.py Main: Flask REST-API
│ │ ├── app_utils.py Helper functions for quantifying geneset similarity
│ │ ├── wsgi.py WSGI production server interface (for use with *gunicorn*)
│ │ │
│ │ ├── upload.py Main: GMTx upload + API upload
│ │ ├── db_utils.py GeMS database initialisation logic
│ │ ├── map_utils.py Use NCBI collections to infer gene IDs and symbols
│ │ ├── gmtx_utils.py Helper functions for parsing GMTx files
│ │ │
│ │ └── README.md Documentation for the REST-API
│ │
│ ├── ncbi_gene_mapper Deprecated: NCBI Gene and Homologene to MongoDB
│ └── x_to_gmtx_converter Conversion to GMTx files
└── ...
cp start_podman.sh start_podman_localhost.sh
Edit start_podman_localhost.sh and change ENV variables to configure the connection to MongoDB instance
Then run:
bash start_podman_localhost.sh
db.getCollection('GeMS_set2set').aggregate([{
$graphLookup: {
from: 'GeMS_set2set',
startWith: '$setId',
connectFromField: 'setId',
connectToField: 'parentId',
as: 'children',
maxDepth: 10,
depthField: 'depth'
}
}])