Public version of bksnake - biokit snakemake - bulk RNASeq Snakemake workflow
- Introduction
- Overview of the analysis workflow
- Requirements
- Preparation
- Configuration
- Usage
- Output
- Contributing
- License
Introduction (top)
bksnake is a Snakemake (Moelder et al., 2021) workflow for bulk RNASeq data analysis. It uses STAR aligner (Dobin et al., 2012) for read mapping and FeatureCounts from the Subread package (Liao et al., 2014) for gene quantification. Reference genomes with RefSeq, Ensembl, and Gencode gene annotations are available for several species such as Homo sapiens (hg38, chm13), Mus musculus (mm10, mm39), Rattus norvegicus (rn6, rn7), Macaca mulatta (Mmul_10), Macaca fascicularis (mfa5, mfa6), Chlorocebus sabaeus (Vero_WHO_p1.0), Sus scrofa (ss11), and Oryctolagus cuniculus (oc2). The generation of these reference genomes and annotation files is documented in a separate repository on github (see refsnake). Data quality and RNASeq metrics are determined using FastQC (Andrews et al.), MultiQC (Ewels et al., 2015), and Picard tools (Broad Institute). In addition, diagnostic plots for data quality assessment such as BioQC tissue heterogeneity (Zhang et al., 2017) or Principal Component Analysis are provided in an HTML report generated by using the Snakemake optional parameter --report. Optionally, genome coverage files (BigWig) and read alignment files (BAM/CRAM) can be generated as well. In addition, cross-checking sample matching using SNP fingerprints and Picard tools (Broad Institute) can be utilized. Input read trimming with Cutadapt (Martin, 2010) and generation of unmapped reads are also available.
The pipeline can be launched via a helper tool, run.py, or directly with Snakemake for users familiar with the workflow tool. All parameters for the pipeline are specified within a configuration yaml file or explicitly on the command line when using run.py. All input data, i.e. input fastq files, a human-readable tab-delimited file describing the samples, and the reference genome and STAR index files, must be available to the pipeline in a local data folder. To run the pipeline, Snakemake and Singularity must be installed and pre-configured. All software tools used by the pipeline are pulled from public Singularity or Docker image repositories. It is recommended to run the pipeline in a high-performance cluster environment.
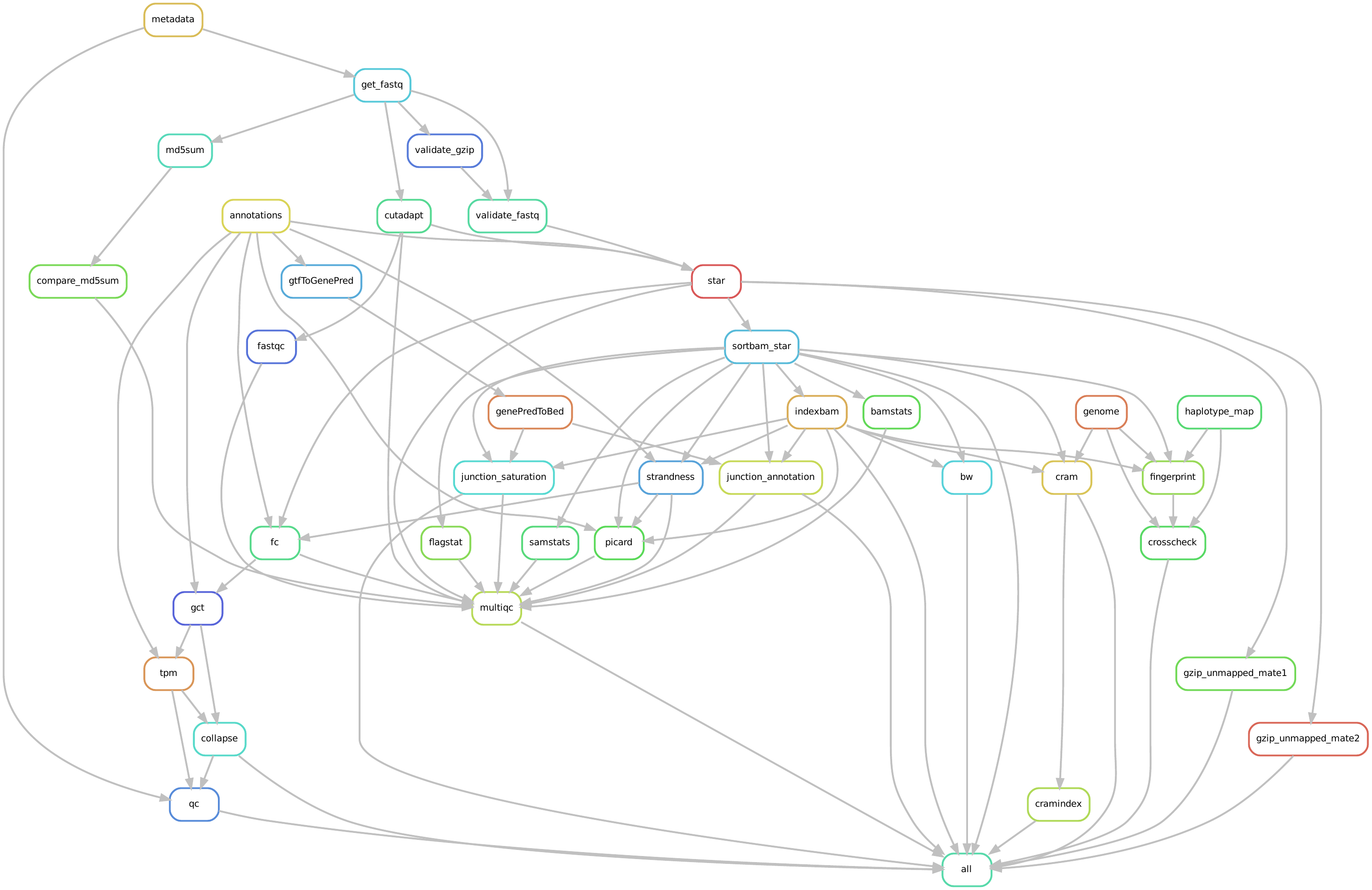
Overview of the analysis workflow (top)
Directed acyclic graph of a representative analysis workflow.
Requirements (top)
This workflow requires the following tools in the system path
- Singularity
- Snakemake (version 7.16.1)
- git
In order to pull Singularity images from GitHub package registry one needs to specify username and GitHub read package token:
export SINGULARITY_DOCKER_USERNAME=<username>
export SINGULARITY_DOCKER_PASSWORD=<github read package token>Preparation (top)
Download workflow (top)
Clone this repository to your local working directory
git clone https://github.com/bedapub/bksnake.gitReference genome (top)
Download prepared reference genome annotation files from Zenodo into a genome "root" directory and "untar" the downloaded files there. At the end, create a symbolic link "hg38" pointing to the data folder for the human genome hg38 files.
genome_dir=<genome root directory>
mkdir -p $genome_dir
cd $genome_dir
wget https://zenodo.org/record/8017214/files/file.dat
tar –xvzf file.dat
ln -s genomes_2022-07-15_hg38 hg38
In folder genomes_2022-07-15_hg38, or hg38, are several subfolders containing fasta and gtf files for RefSeq and Ensembl annotations as well as index files for the STAR aligner version 2.7.10b.
<genome root directory>/hg38
├── fasta
├── gtf
│ ├── ensembl
│ └── refseq
├── star_2.7.10b
Specify the genome "root" directory in the pipeline configuration file (config/config.yaml), parameter genome_dir (see below).
Other species (top)
Reference genomes of other species can be generated, for example, with the refsnake Snakemake workflow.
Metadata (top)
The main input to the workflow is a tab-delimited text file containing metadata information about all samples.
See an example file, resources/test-data/metadata.txt, in the previously cloned pipeline directory.
The metadata file is composed of one header line and sample information on subsequent lines, one sample per line, organized by several columns as follows:
#ID: unique sample labelGROUP: name of sample conditionFASTQ1: name of forward read fastq file (mate 1, R1)FASTQ2: name of reverse read fastq file (mate 2, R2)Raw: path to the folder containing the input fastq filesOrganism: Human, Rat, Mouse, or Pig
Note that columns #ID and GROUP may not contain white-spaces and other special characters.
It is possible to add additional columns, for example, to describe additional experiment parameters or specimen information. But they are not used by the pipeline further.
Example (top)
| #ID | GROUP | FASTQ1 | FASTQ2 | Raw | Organism |
|---|---|---|---|---|---|
| GSM5362225 | HOXB9-KO | 10k_SRR14749833_1.fastq.gz | 10k_SRR14749833_2.fastq.gz | resources/test-data/fastq | Human |
| GSM5362224 | HOXB9-T133A | 10k_SRR14749832_1.fastq.gz | 10k_SRR14749832_2.fastq.gz | resources/test-data/fastq | Human |
| GSM5362223 | HOXB9 | 10k_SRR14749831_1.fastq.gz | 10k_SRR14749831_2.fastq.gz | resources/test-data/fastq | Human |
Configuration (top)
The workflow requires several parameters to be configured, most of which can be set through a yaml configuration file.
A template file named config.yaml is provided in the config directory.
It's recommended to create a local copy of the template and make modifications there.
Note that many of these parameters can also be specified through the wrapper script run.py, as explained in the next section.
Parameters specified on the command line through the wrapper script run.py will overwrite parameters set in the configuration file.
To learn about all possible parameters, execute:
python run.py --helpIt is important to specify the following parameters in the config.yaml
- genome root and sub- directory:
genome_dir: VALUE - singularity images directory:
singularity: prefix: VALUE - snakemake path:
snakemake: path: VALUE - sample metadata file:
metadata: file: VALUEandmetadata: group_name: VALUE - sequencing library:
library: type: VALUEandlibrary: strand: VALUE
Usage (top)
Run on a cluster with LSF scheduler, up to 100 jobs in parallel
python run.py --config <path to config.yaml> --jobs 100Run locally, using up to 12 cores
python run.py --config <path to config.yaml> --cores 12Run the pipeline with the test data set and specify four optional parameters on the command line
python run.py \
--config config/config.yaml \
--metadata-file resources/test-data/metadata.txt \
--snakemake-path=<path to snakemake> \
--genome-dir <genome root directory> \
--outdir test-data_output \
--jobs 50By this, sample metadata, in particular the path for the raw input data, i.e. "fastq" files, are given by the input metadata file located at resources/test-data/metadata.txt.
The test data set consists of three sub samples fastq files from A549 cell line samples from GEO study GSM5362223.
By the command above, the Snakemake jobs will be submitted to the cluster queue. A maximum of 50 jobs will be processed simultaneously.
All output will be written to a new folder named test-data_output.
All other pipeline configuration parameters will be used from the default config template file, config/config.yaml.
Output (top)
Folder structure of a typical workflow run (* = optional output).
├── annot genome annotation files
├── bam* read mappings in BAM format (optional)
├── bw* read coverage BigWig files (optional)
├── config.yaml workflow configuration file
├── cram* read mappings in CRAM format (optional)
├── cutadapt* Cudapapt output (optional)
├── fastq* copy of input reads (optional)
├── fastqc FASTQC output files
├── fc FeatureCounts output files
├── gct gene counts and normalized gene counts in GCT file format for RefSeq/Ensembl/Gencode annotations
├── log log and output files from the tools used
├── metrics* CrosscheckFingerprints file from Picard tools for human data (optional)
├── multiqc_data MultiQC data files
├── multiqc_report_ensembl.html MultiQC report for Ensembl annotations
├── multiqc_report_gencode.html MultiQC report for Gencode annotations (human and mouse)
├── multiqc_report_refseq.html MultiQC report for RefSeq annotations
├── qc some QC plots, e.g. PCA or BioQC
├── report.html Snakemake report, containing workflow and QC metrics
├── rulegraph.pdf workflow DAG in pdf format
├── rulegraph.png workflow DAG in png format
├── samples.txt sample metadata in tab-delimited file
└── unmapped* unmapped reads in FASTQ file format (optional)
Explanations (top)
Contains copies of reference genome and annotations files (i.e. FASTA, GTF, etc)
In addition, sample metadata in tab-delimited text file, phenoData.meta and in cls format phenoData.cls.
Contains all aligned reads in BAM file. Only generated if the parameter keep_bam_files is True in the pipeline configuration.
Read coverage files in BigWig format. May be used for graphical visualization of the genome coverage by external tools such as JBrowse or IGV. Only generated if the parameter generate_bw_files is True in the pipeline configuration.
Output files from the sequencing read trimming tool Cutadapt. Only generated if the parameter cutadapt: run is True in the pipeline configuration.
All configuration parameters stored in a single yaml file.
Contains all aligned reads in CRAM file. Only generated if the parameter generate_cram_files is True in the pipeline configuration.
Contains a copy of the input FASTQ files. Only generated if the parameter keep_fastq_files is True in the pipeline configuration.
Contains all output files from the read quality control tool FastQC.
Intermediate output files, summary and compressed counts file, from the gene quantification step by using FeatureCounts tool from the Subreads package. Parameters for FeatureCounts can be specified via the input configuration file.
Contains - if available - RefSeq, Ensembl, or Gencode (<db>) annotated gene counts, normalized counts, log2-transformed counts in GCT file format.
<db>_count.gct: RefSeq gene counts<db>_count_collapsed.gct: RefSeq gene counts collapsed to human orthologous gene symbols (usingresources/geneids.chip)<db>_tpm.gct: normalized RefSeq transcript per million mapped reads (tpm)<db>_tpm_collapsed.gct: human orthologs of normalized counts (tpm)<db>_log2tpm.gct: log2-transformed normalized RefSeq transcript per million mapped reads (tpm)
Contains several log files from analysis tools used by the pipeline. Mainly used for debugging purposes.
There is an optional rule for cross-checking human sample matching using SNP fingerprints and the Picard tool (BroadPicard-Tool).
A haplotype map is a collection of "blocks" of SNPs that are in tight linkage with SNPs of the same block and low linkage with SNPs of different blocks (see Javed et al., 2020). Here, we utilize fingerprint maps from naumanjaved on github. In order to use these features, set the config file parameter crosscheck_fingerprints: True.
Output files from the MultiQC tool with RefSeq, Ensembl, Gencode gene annotations (e.g. for Picard RNASeq metrics).
HTML summary report from the MultiQC tool based on genome annotations from <db> where <db> is refseq, ensembl, gencode (if available).
Plots for quality control purposes, using genome annotations from <db>.
<db>_bioQC.pdf: Heatmap representation of the BioQC enrichment scores for detecting detecting such tissue heterogeneity (Zhang et al. 2017)<db>_bioQC_thr2.txt: BioQC enrichment scores above threshold 2<db>_bioQC.txt: All BioQC enrichment scores<db>_log2tpm_pca.pdf: Plot of the main components from the principal component analysis on the basis of the log2-transformed normalized gene counts (log2tpm).<db>_log2tpm_pca.txt: Coordinates of the components from the principal component analysis on the basis of the log2-transformed normalized gene counts (log2tpm).
Graphical representation of the entire workflow. A directed acyclic graph, DAG, generated by Snakemake
Tab-delimited, human readable text file containing study and sample metadata in tabular form (one sample per line).
Contains unmapped sequencing reads in FASTQ files. Only generated if the parameter generate_unmapped is True in the pipeline configuration.
Contributing (top)
Any contribution, feedback and bug report are highly welcome. For major changes, please open an issue first to discuss what you would like to change. Thank you!