This code accompanies the paper: Receptor endocytosis orchestrates the spatiotemporal bias in β-arrestin signaling.
It contains the following files:
- code/model_vectorised.py: running the simulations
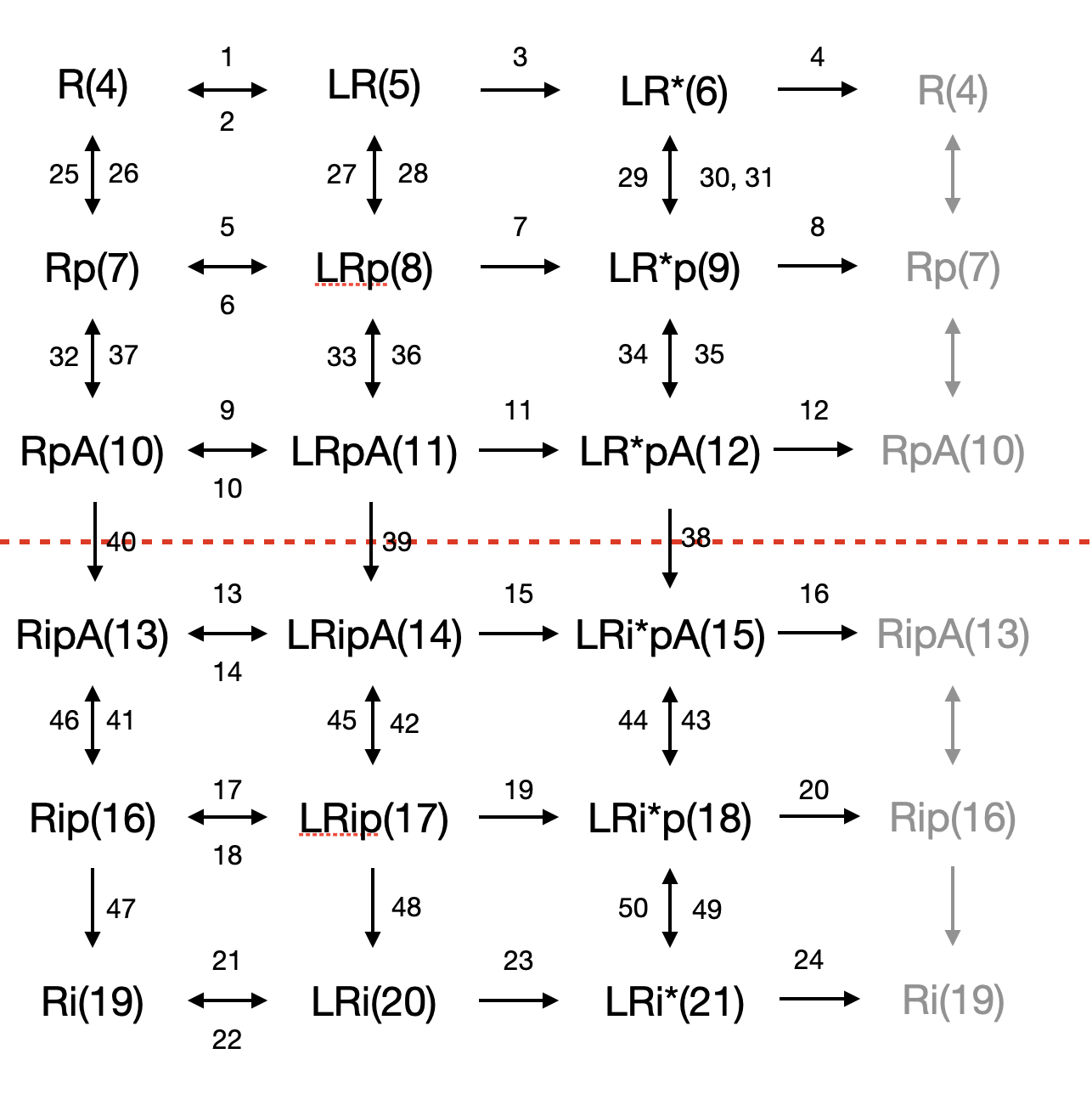
- model/toth_szalai_gpcr.txt: the concentraions and reaction constants for ODE models
- code/00-model_tutorial.ipynb: some tutorial
- code/01-run_models.ipynb: code to run the simulations in the paper
- code/02-figures.ipynb: code to reproduce the figures of the paper