A PyTorch implementation of Semi-Supervised Graph Classification: A Hierarchical Graph Perspective (WWW 2019)
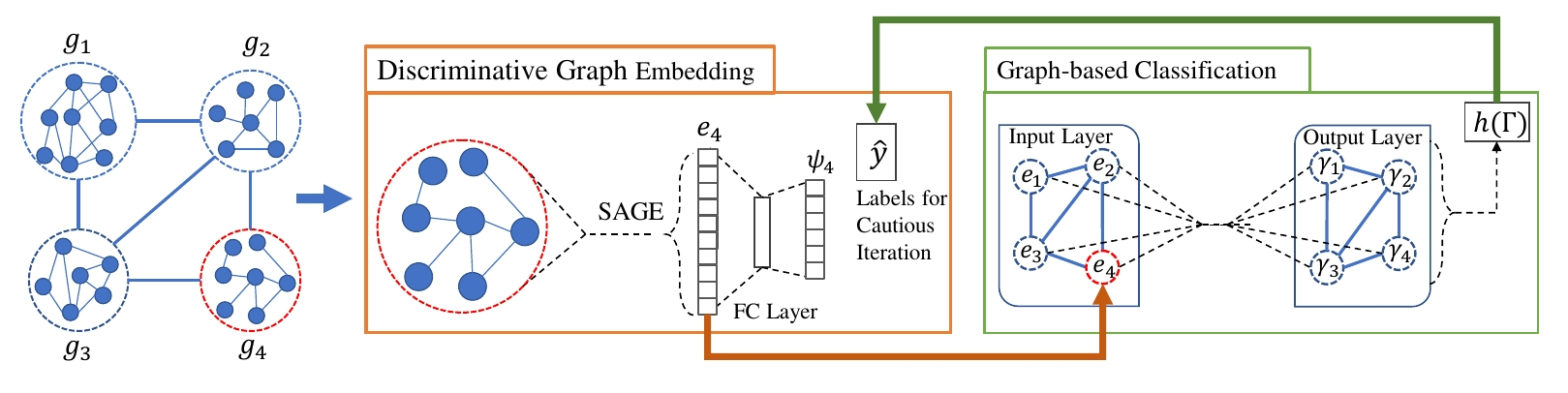
Node classification and graph classification are two graph learning problems that predict the class label of a node and the class label of a graph respectively. A node of a graph usually represents a real-world entity, e.g., a user in a social network, or a protein in a protein-protein interaction network. In this work, we consider a more challenging but practically useful setting, in which a node itself is a graph instance. This leads to a hierarchical graph perspective which arises in many domains such as social network, biological network and document collection. For example, in a social network, a group of people with shared interests forms a user group, whereas a number of user groups are interconnected via interactions or common members. We study the node classification problem in the hierarchical graph where a `node' is a graph instance, e.g., a user group in the above example. As labels are usually limited in real-world data, we design two novel semi-supervised solutions named Semi-supervised graph classification via Cautious/Active Iteration (or SEAL-C/AI in short). SEAL-C/AI adopt an iterative framework that takes turns to build or update two classifiers, one working at the graph instance level and the other at the hierarchical graph level. To simplify the representation of the hierarchical graph, we propose a novel supervised, self-attentive graph embedding method called SAGE, which embeds graph instances of arbitrary size into fixed-length vectors. Through experiments on synthetic data and Tencent QQ group data, we demonstrate that SEAL-C/AI not only outperform competing methods by a significant margin in terms of accuracy/Macro-F1, but also generate meaningful interpretations of the learned representations.
This repository provides a PyTorch implementation of SEAL-CI as described in the paper:
Semi-Supervised Graph Classification: A Hierarchical Graph Perspective. Jia Li, Yu Rong, Hong Cheng, Helen Meng, Wenbing Huang, Junzhou Huang. WWW, 2019. [Paper]
A TensorFlow implementatio of the model is available [here].
The codebase is implemented in Python 3.5.2. package versions used for development are just below.
networkx 2.4
tqdm 4.28.1
numpy 1.15.4
pandas 0.23.4
texttable 1.5.0
scipy 1.1.0
argparse 1.1.0
torch 1.1.0
torch-scatter 1.4.0
torch-sparse 0.4.3
torch-cluster 1.4.5
torch-geometric 1.3.2
torchvision 0.3.0
The code takes graphs for training from an input folder where each graph is stored as a JSON. Graphs used for testing are also stored as JSON files. Every node id and node label has to be indexed from 0. Keys of dictionaries are stored strings in order to make JSON serialization possible.
The graphs file has to be unzipped in the input folder.
Every JSON file has the following key-value structure:
{"edges": [[0, 1],[1, 2],[2, 3],[3, 4]],
"features": {"0": ["A","B"], "1": ["B","K"], "2": ["C","F","A"], "3": ["A","B"], "4": ["B"]},
"label": "A"}The edges key has an edge list value which descibes the connectivity structure. The features key has features for each node which are stored as a dictionary -- within this nested dictionary features are list values, node identifiers are keys. The label key has a value which is the class membership.
The hierarchical graph is stored as an edge list, where graph identifiers integers are the node identifiers. Finally, node pairs are separated by commas in the comma separated values file. This edge list file has a header.
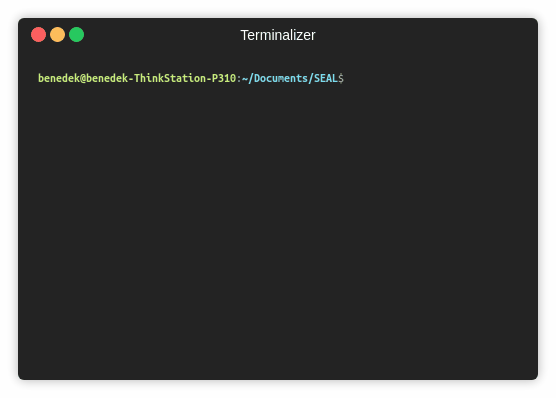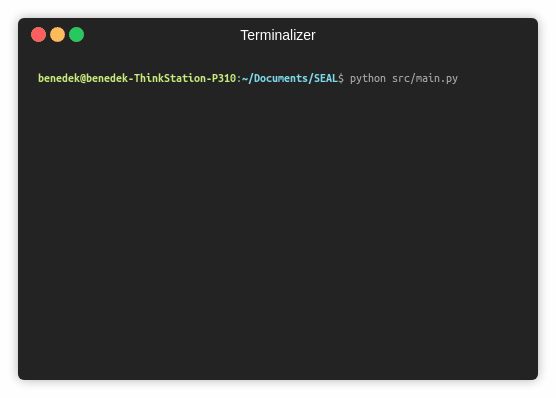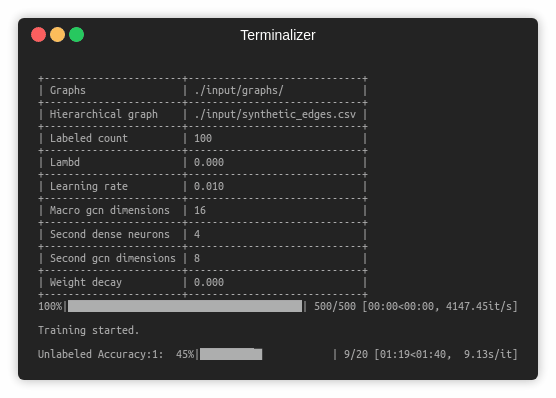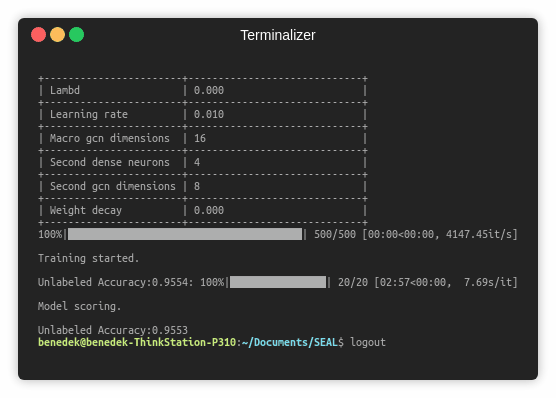
Training a SEAL-CI model is handled by the src/main.py script which provides the following command line arguments.
--graphs STR Training graphs folder. Default is `input/graphs/`.
--hierarchical-graph STR Macro level graph. Default is `input/synthetic_edges.csv`.
--epochs INT Number of epochs. Default is 10.
--budget INT Nodes to be added. Default is 20.
--labeled-count INT Number of labeled instances. Default is 100.
--first-gcn-dimensions INT Graph level GCN 1st filters. Default is 16.
--second-gcn-dimensions INT Graph level GCN 2nd filters. Default is 8.
--first-dense-neurons INT SAGE aggregator neurons. Default is 16.
--second-dense-neurons INT SAGE attention neurons. Default is 4.
--macro-gcn-dimensions INT Macro level GCN neurons. Default is 16.
--weight-decay FLOAT Weight decay of Adam. Defatul is 5*10^-5.
--gamma FLOAT Regularization parameter. Default is 10^-5.
--learning-rate FLOAT Adam learning rate. Default is 0.01.
The following commands learn a model and score on the unlabaled instances. Training a model on the default dataset:
python src/main.py
Training each SEAL-CI model for a 100 epochs.
python src/main.py --epochs 100
Changing the budget size.
python src/main.py --budget 200