Welcome to the repository from the iGEM.CNPEM Design League team, focused on the Mathematical Modeling of the B.A.R.B.I.E. project!
Bioengineered Approach for Removal of microplastics through Bioremediation and Innovative Electromagnetics — or
B.A.R.B.I.E.— proposes an innovative treatment stage in Water Plants (WTPs). Our approach is implemented after classical filtration with a bio-based process specifically designed to detect and capture Microplastics (MPs) present in the water.
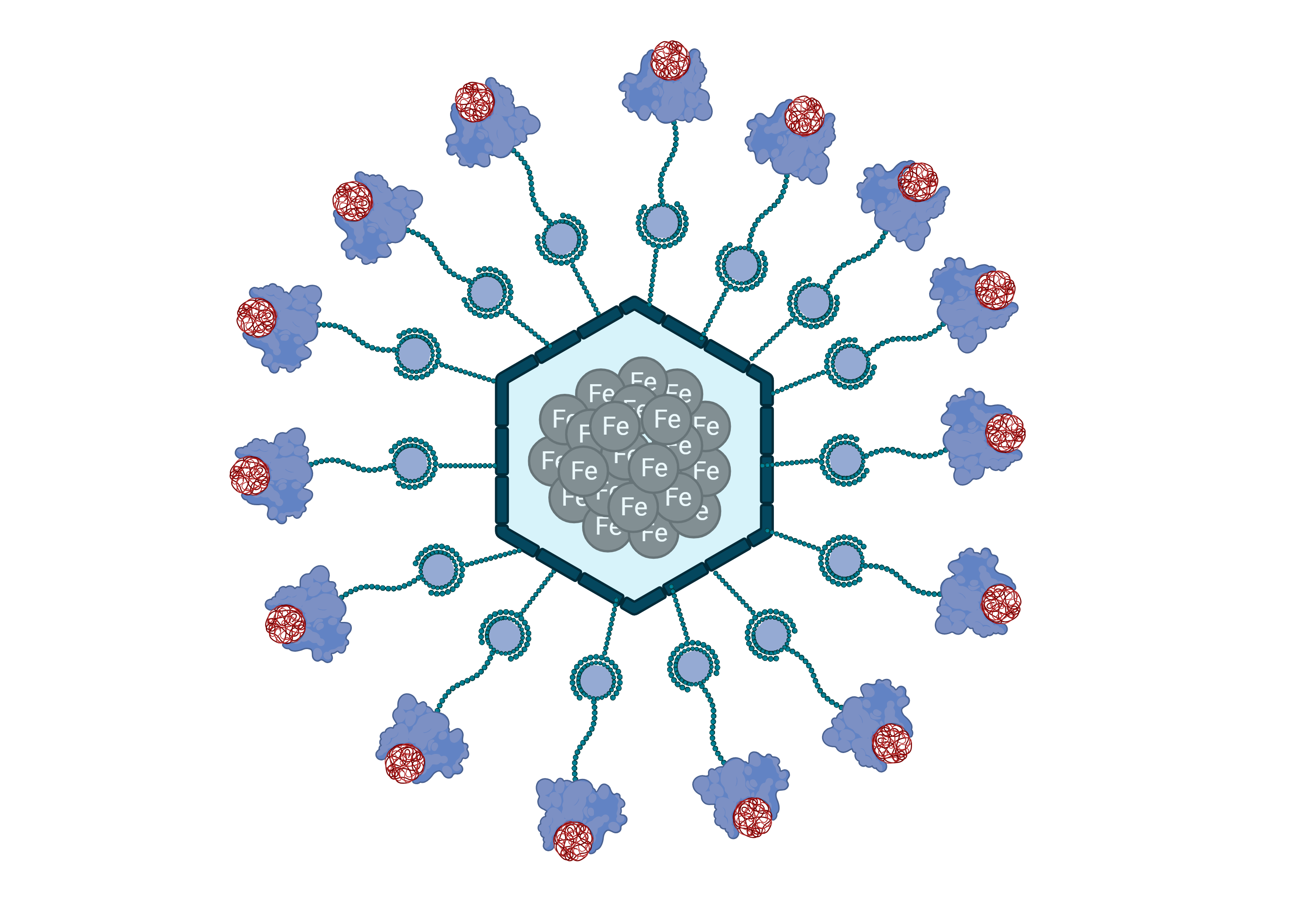
The ongoing strategy combines magnetic nanoparticles (MNPs) with biological molecules, such as Plastic Binding Proteins (PBP). Thus, the BaCBM2 protein (Bacillus anthraci Carbohydrate Binding Molecule, family 2), which binds to environmentally relevant plastics, moonlights as the key component of B.A.R.B.I.E.
The proposed strategy includes adding BaCBM2 proteins linked to MNPs, to the treated water. The MNPs, when directed by controlled external magnetic fields, will allow the removal of the nanoparticle-BaCBM2-MP complex. Concomitantly, we propose the construction of a sensor to detect the presence of MPs in water samples.
This sensor uses impedance spectroscopy and machine learning techniques that allow the individual characterization of particles based on their electrical properties, simultaneously identifying the material and size of the particles.
In this repository, you will find data and code related to the metabolic modeling of our chassis, aiming to optimize the production and purification of the proteins used in our complex, including:
EncapsulinPlastic Binding Protein (PBP)SPYs
For this, at this point, we made a model to describe the cells growth, and estimate the proteins productions. The model was presented in the notebook "cells_modelling.ipynb".
Who are the students behind this Mathematical Modeling iGEM.CNPEM project? Well, allow us to introduce ourselves:
- 👋 We are Gabriel Pereira, Isabela Beneti, Pedro Zanineli and Gustavo Beneti.
- 📕 We are students from the The Brazilian Center for Research in Energy and Materials (CNPEM).
- 👨🔬 CNPEM hosts 4 national laboratories, including SIRIUS (Brazilian particle accelerator) and LNBR (Brazilian National Laboratory for BioRenewables).
- 💖 CNPEM has its own Design League team, iGEM.CNPEM, which this year created the B.A.R.B.I.E. project
- 👩💻 This repository aims to store all the data and code developed in the mathematical modeling branch of the iGEM.CNPEM team.




Without the help of our mentors, we wouldn't have been able to reach where we are now! Therefore, we thank the following individuals who contributed to this project:
 Vinícius Wasques (Professor) |
 João Eduardo Levandoski (Mentor) |
 Alessandro Mourato (Técnico) |