ggDiagnose
This package is geared towards data scientists, statistics students and the broader statistics community who increasingly use ggplot2 and tidyverse tools to explore and visualize their data. In this package we provide functions that create ggplot2 based visualizations that can replace the base plot function to diagnose and visualize R objects. Specifically, this package aims to provide visualation tools when these objects are model objects and other non-traditional (read: non-data frame) based objects.
For examples of visuals see the example section.
Installation
To install this function, just do the following:
library(devtools)
devtools::install_github("benjaminleroy/ggDiagnose")This package requires very few packages; if dependencies are required for a specific object's functions, the user is prompted to install such packages.
Future of the Package
This package was envisioned as a package that would naturally grow to meet the needs of the users. As such, please (1) feel free to create an issue to request ggDiagnose functionality for R objects and (2) develop these missing modules and submit a merge request to improve the package.
Examples
Links to examples:
ggDiagnose
ggDiagnose.lmggDiagnose.glmnetggDiagnose.cv.glmnetggDiagnose.GamggDiagnose.treeggDiagnose.matrix- ... more to come ...
dfCompile
dfCompile.lmdfCompile.glmnetdfCompile.cv.glmnetdfCompile.GamdfCompile.treedfCompile.matrix- ... more to come ...
We also provide a .rmd file with the same examples runable in Rstudio, which can be found here, or by just heading to the folder labeled rmarkdown_examples and downloading the ggDiagnose_examples.rmd file.
ggDiagnose
ggDiagnose.lm
This example is for an lm object, function works for glm and rlm objects as well.
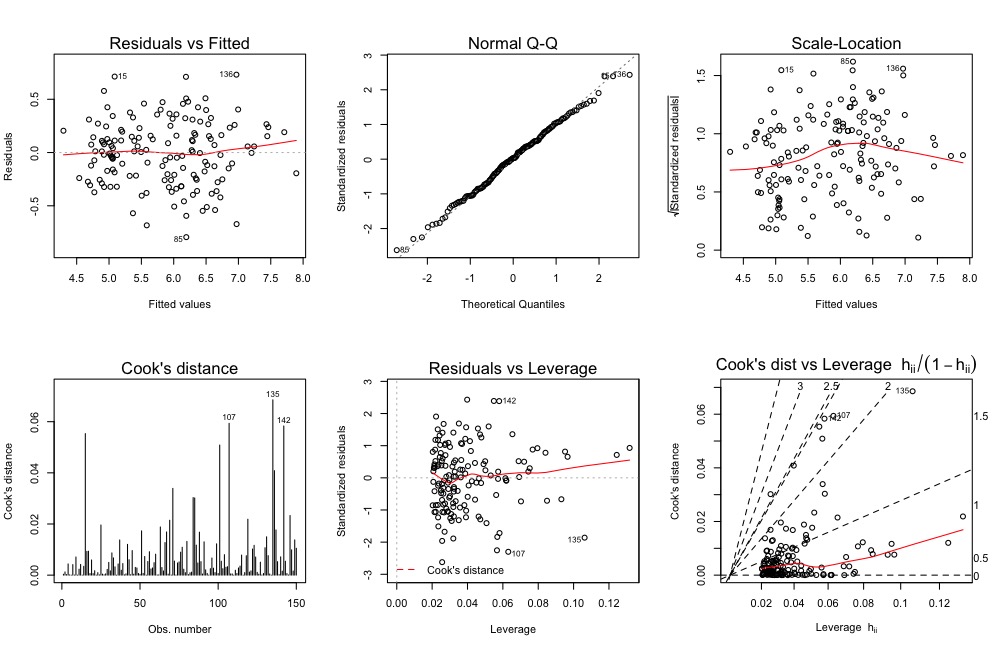
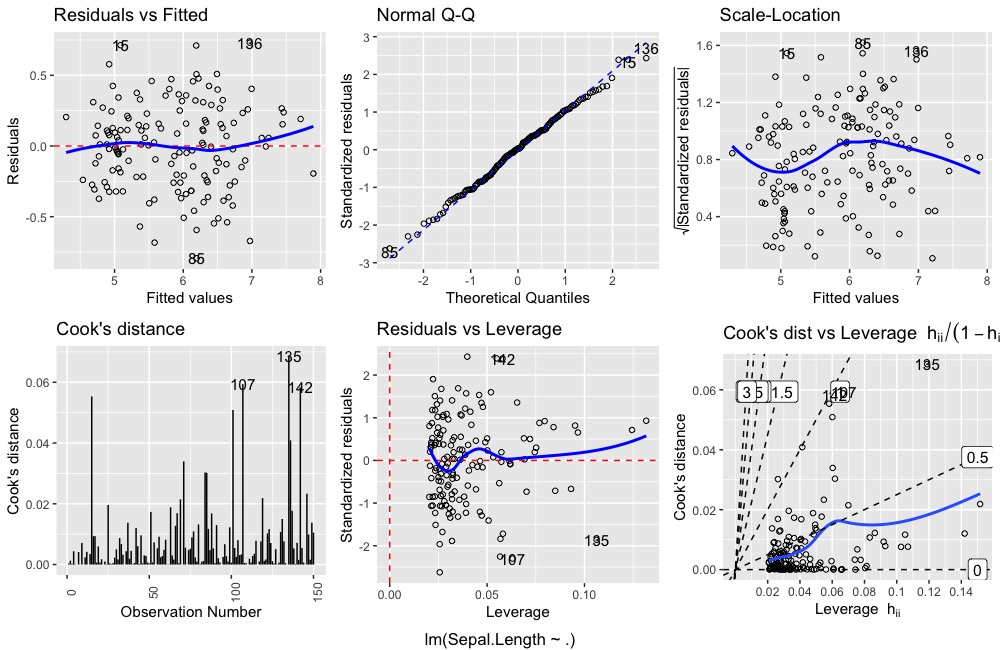
lm.object <- lm(Sepal.Length ~., data = iris)The original visualization:
par(mfrow = c(2,3))
plot(lm.object, which = 1:6)The updated visualization:
ggDiagnose(lm.object, which = 1:6)ggDiagnose.lm allows for similar parameter inputs as plot.lm but also includes additional ones. This may changes as the package evolves.
ggDiagnose.glmnet
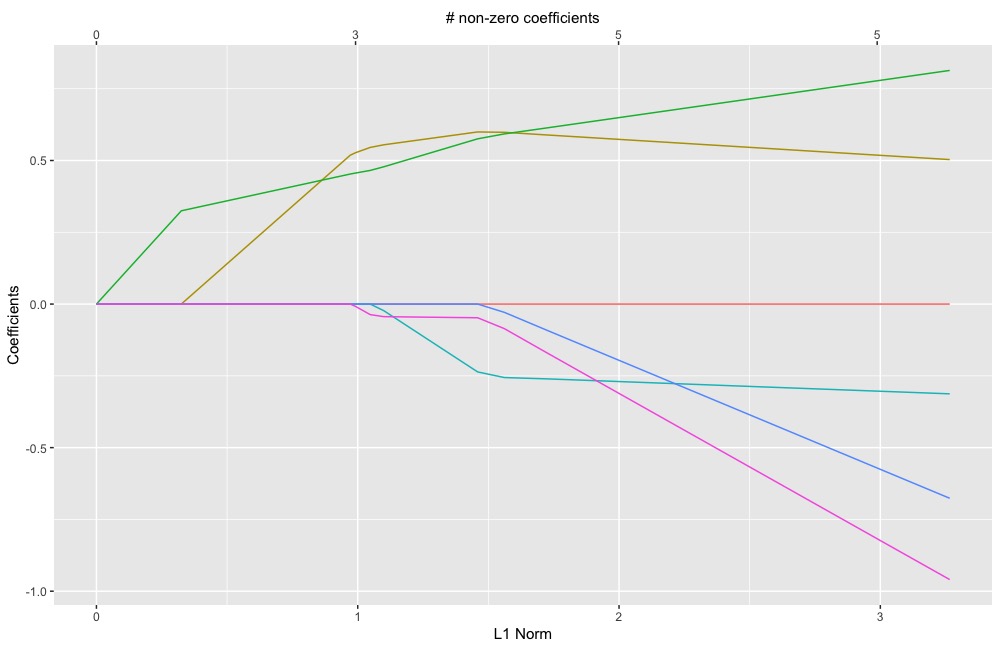
library(glmnet)
glmnet.object <- glmnet(y = iris$Sepal.Length,
x = model.matrix(Sepal.Length~., data = iris))The original visualization:
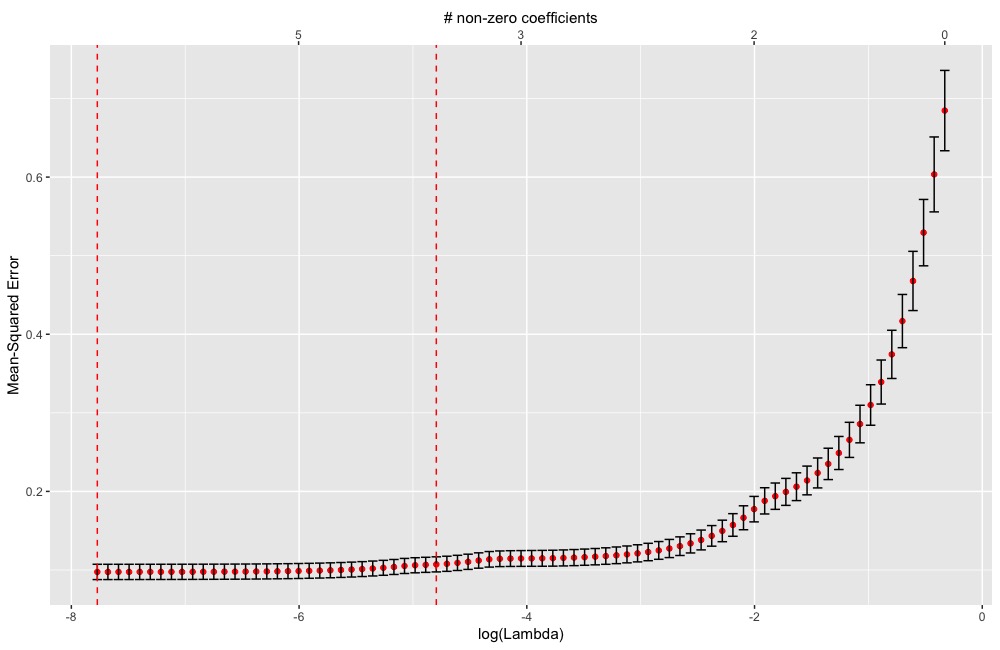
plot(glmnet.object)ggDiagnose(glmnet.object)ggDiagnose.cv.glmnet
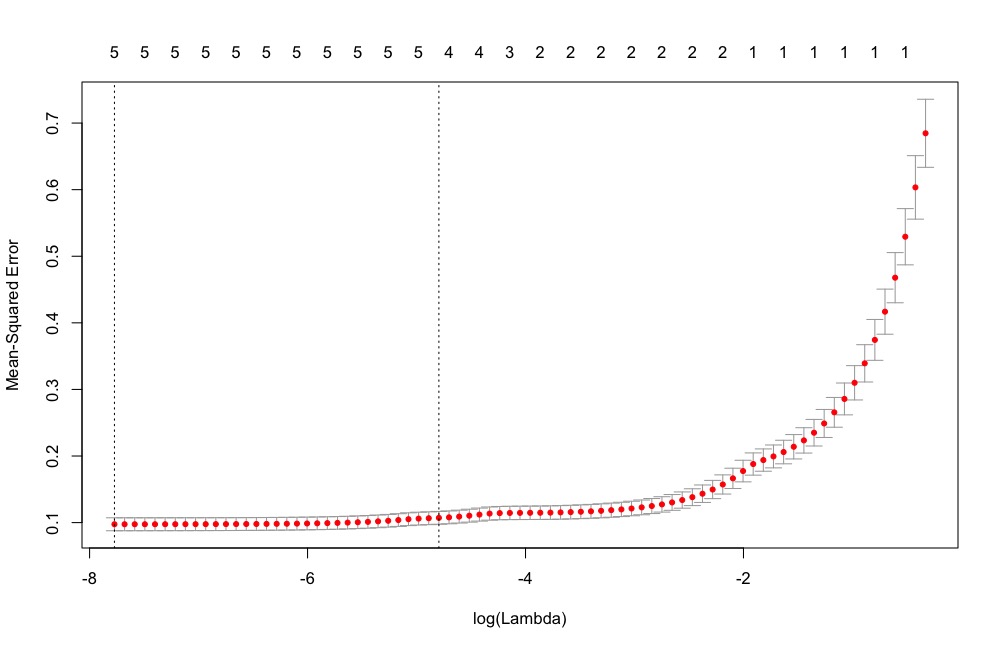
cv.glmnet.object <- cv.glmnet(y = iris$Sepal.Length,
x = model.matrix(Sepal.Length~., data = iris))The original visualization:
plot(cv.glmnet.object)ggDiagnose(cv.glmnet.object)ggDiagnose.Gam
library(gam)
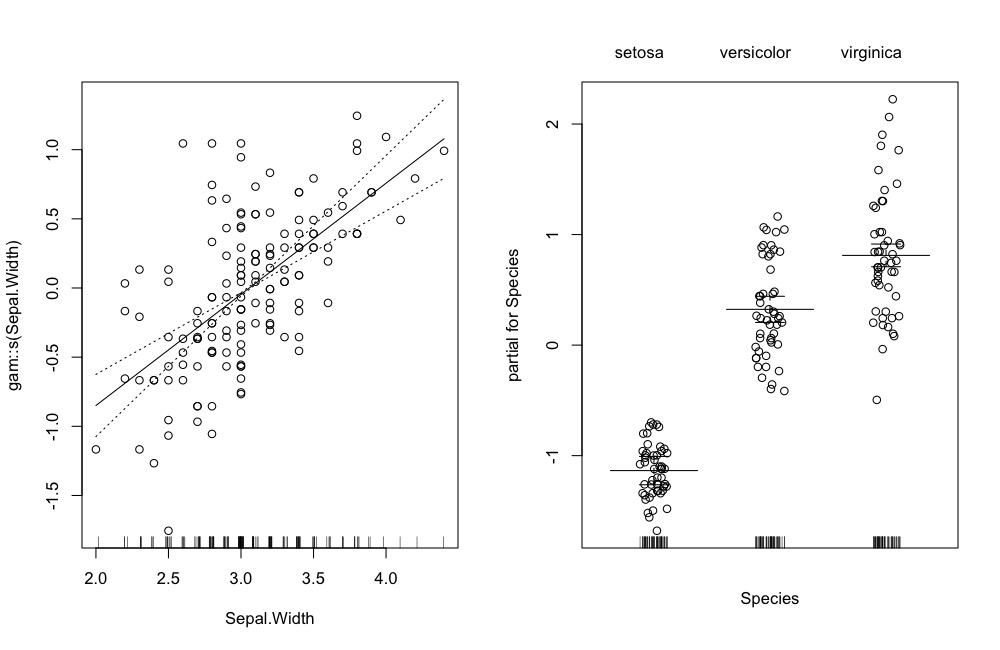
gam.object <- gam::gam(Sepal.Length ~ gam::s(Sepal.Width) + Species,
data = iris)The original visualization:
par(mfrow = c(1,2))
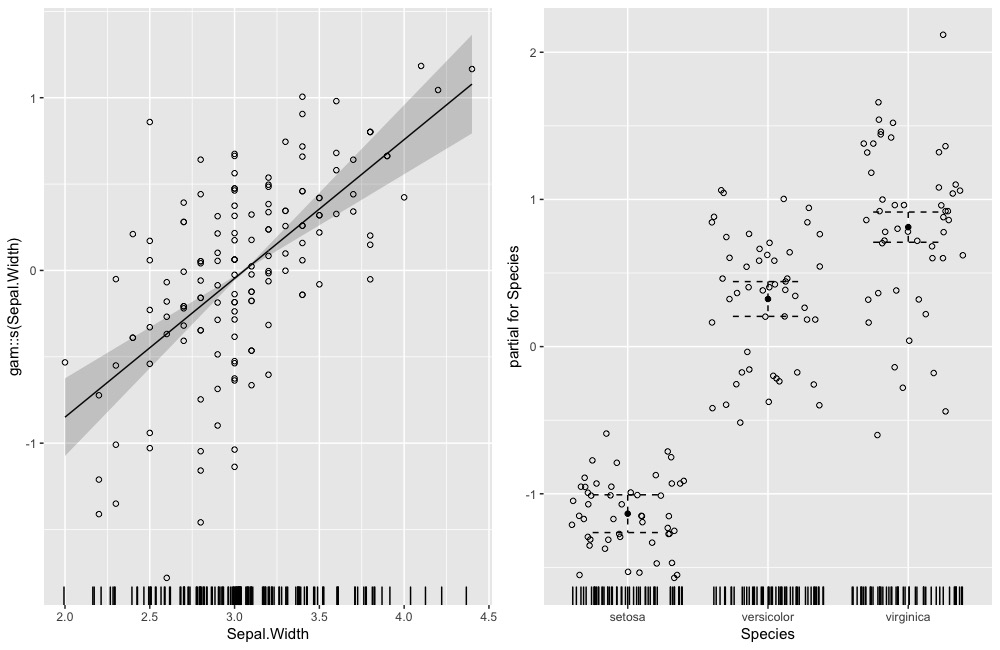
plot(gam.object, se = TRUE, residuals = TRUE)The updated visualization:
ggDiagnose(gam.object, residuals = TRUE) # se = TRUE by default
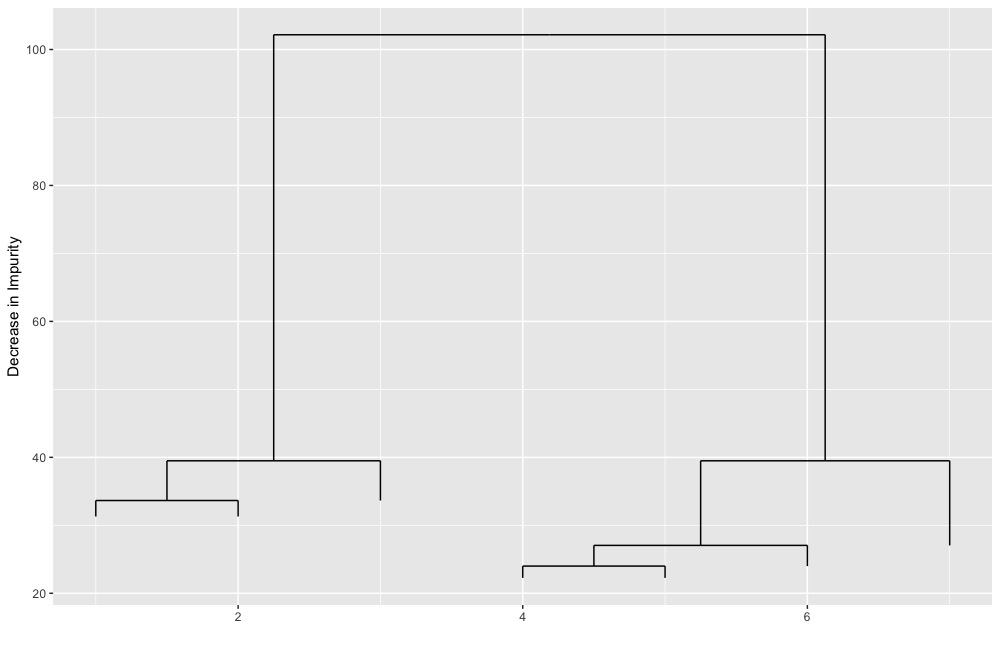
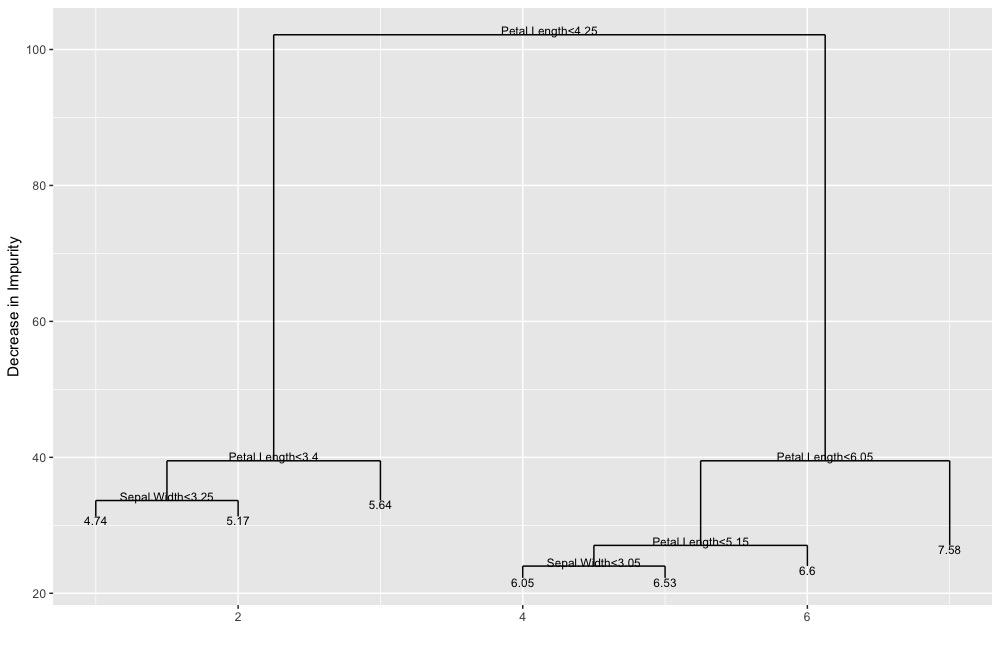
ggDiagnose.tree
Note, for more perfect replication of the base plot function add + ggdendro::theme_dendro() which drops all ggplot background elements.
library(tree)
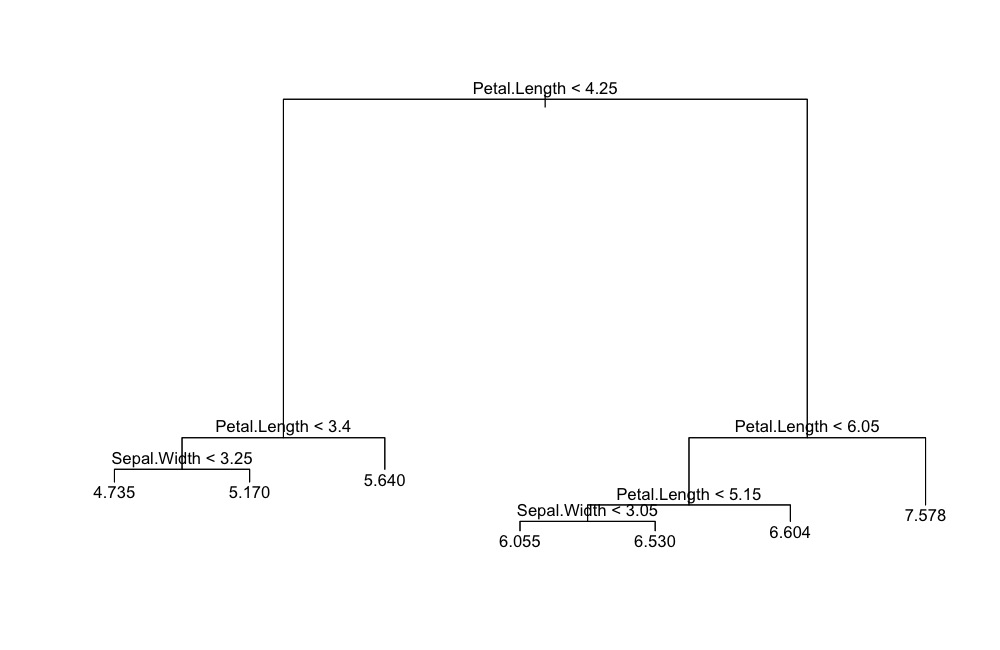
tree.object <- tree(Sepal.Length ~., data = iris)The original visualization:
plot(tree.object)
text(tree.object)The updated visualization (followed by quick improvement):
ggDiagnose(tree.object, split.labels = FALSE)ggDiagnose(tree.object, split.labels = TRUE,
leaf.labels = TRUE)ggDiagnose.matrix
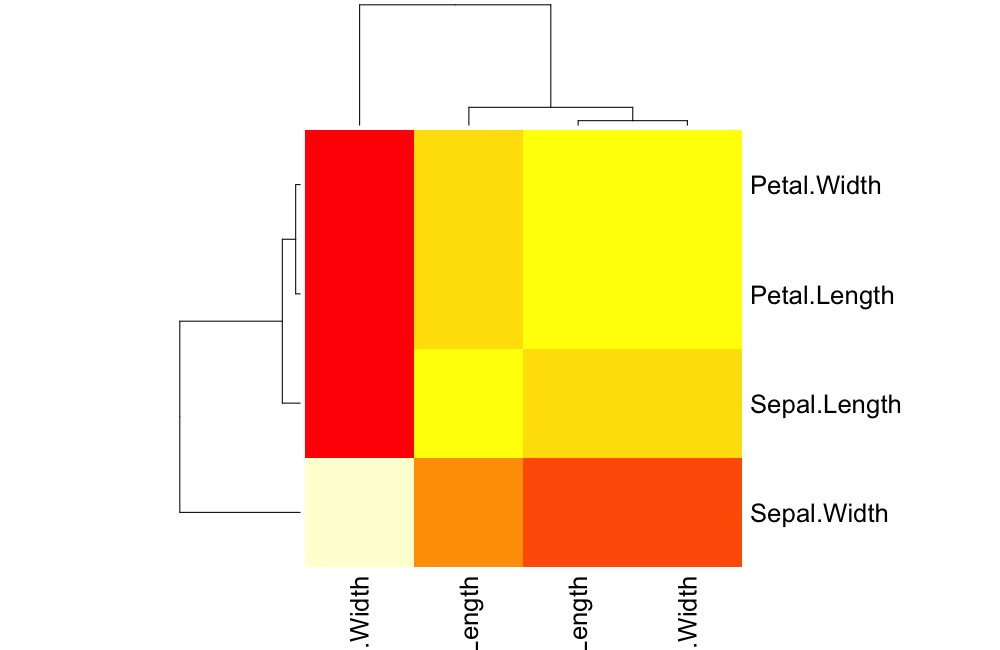
ggDiagnose.matrix actually can create a visualization of a matrix similar to the "base R"'s image or heatmap.
Heatmap visualization
The original visualization using heatmap
data(iris)
iris_c <- iris %>% select(-Species) %>% cor %>% as.matrix
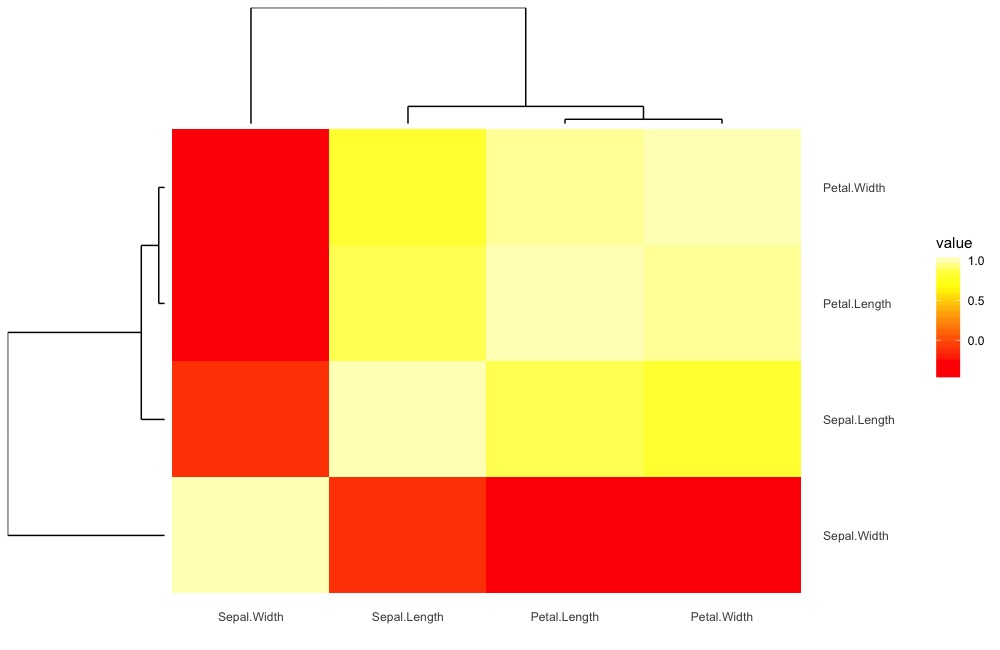
heatmap(iris_c)The updated visualization (note that we did a little manipulation to make the default color scheme match.) We may make this the default within the function later.
ggDiagnose.matrix(iris_c, type = "heatmap",return = T, show.plot = F)$ggout +
scale_fill_gradientn(colours = grDevices::heat.colors(10))Image visualization
The original function image:
myurl = "http://stat.cmu.edu/~bpleroy/images/me.jpg"
data = jpeg::readJPEG(RCurl::getURLContent(myurl))[,,1]
data = t(data)[,rev(1:nrow(data))]image(data, col = grey(seq(0, 1, length = 256)))The updated visualization (note that again we changed the base color to match with the expected output for this image.)
ggDiagnose.matrix(data, type = "image",return =T, show.plot=F)$ggout +
scale_fill_gradient(high = "white",low = "black") dfCompile
dfCompile.lm
> library(dplyr)
> lm.object <- lm(Sepal.Length ~., data = iris)> dfCompile(lm.object) %>% names
[1] "Sepal.Length" "Sepal.Width" "Petal.Length"
[4] "Petal.Width" "Species" ".index"
[7] ".labels.id" ".weights" ".yhat"
[10] ".resid" ".leverage" ".cooksd"
[13] ".weighted.resid" ".std.resid" ".sqrt.abs.resid"
[16] ".pearson.resid" ".std.pearson.resid" ".logit.leverage"
[19] ".ordering.resid" ".ordering.std.resid" ".ordering.cooks"
[22] ".non.extreme.leverage"> dfCompile(lm.object) %>% head(2) # needs package dplyr for "%>%"
Sepal.Length Sepal.Width Petal.Length Petal.Width Species .index .labels.id
1 5.1 3.5 1.4 0.2 setosa 1 1
2 4.9 3.0 1.4 0.2 setosa 2 2
.weights .yhat .resid .leverage .cooksd .weighted.resid
1 1 5.004788 0.09521198 0.02131150 0.0003570856 0.09521198
2 1 4.756844 0.14315645 0.03230694 0.0012517183 0.14315645
.std.resid .sqrt.abs.resid .pearson.resid .std.pearson.resid .logit.leverage
1 0.3136729 0.5600651 0.09521198 0.3136729 0.02177557
2 0.4742964 0.6886918 0.14315645 0.4742964 0.03338553
.ordering.resid .ordering.std.resid .ordering.cooks .non.extreme.leverage
1 115 115 124 TRUE
2 98 98 96 TRUEdfCompile.glmnet
> library(glmnet)
> glmnet.object <- glmnet(y = iris$Sepal.Length,
x = model.matrix(Sepal.Length~., data = iris))> dfCompile(glmnet.object) %>% names # needs package dplyr for "%>%"
[1] ".log.lambda" "variable" "beta.value" ".norm"
[5] ".dev" ".number.non.zero"> dfCompile(glmnet.object) %>% head(2) # needs package dplyr for "%>%"
.log.lambda variable beta.value .norm .dev .number.non.zero
1 -0.3292550 X.Intercept. 0 0.00000000 0.0000000 0
2 -0.4222888 X.Intercept. 0 0.03632753 0.1290269 1dfCompile.cv.glmnet
> library(glmnet)
> cv.glmnet.object <- cv.glmnet(y = iris$Sepal.Length,
x = model.matrix(Sepal.Length~., data = iris))> dfCompile(cv.glmnet.object) %>% names # needs package dplyr for "%>%"
[1] "cross.validated.error" "cross.validation.upper.error"
[3] "cross.validation.lower.error" "number.non.zero"
[5] ".log.lambda"> dfCompile(cv.glmnet.object) %>% head(2) # needs package dplyr for "%>%"
cross.validated.error cross.validation.upper.error
s0 0.6840064 0.7340895
s1 0.6009296 0.6487392
cross.validation.lower.error number.non.zero .log.lambda
s0 0.6339234 0 -0.3292550
s1 0.5531200 1 -0.4222888dfCompile.Gam
> library(gam)
> gam.object <- gam::gam(Sepal.Length ~ gam::s(Sepal.Width) + Species,
data = iris)> dfCompile(gam.object) %>% names # needs package dplyr for "%>%"
[1] "Sepal.Length"
[2] "Sepal.Width"
[3] "Petal.Length"
[4] "Petal.Width"
[5] "Species"
[6] ".resid"
[7] ".smooth.gam.s.Sepal.Width."
[8] ".smooth.Species"
[9] ".se.smooth.gam.s.Sepal.Width..upper"
[10] ".se.smooth.Species.upper"
[11] ".se.smooth.gam.s.Sepal.Width..lower"
[12] ".se.smooth.Species.lower" > dfCompile(gam.object) %>% head(2) # needs package dplyr for "%>%"
Sepal.Length Sepal.Width Petal.Length Petal.Width Species .resid
1 5.1 3.5 1.4 0.2 setosa 0.03614362
2 4.9 3.0 1.4 0.2 setosa 0.23792407
.smooth.gam.s.Sepal.Width. .smooth.Species
1 0.35570963 -1.135187
2 -0.04607082 -1.135187
.se.smooth.gam.s.Sepal.Width..upper .se.smooth.Species.upper
1 0.44985507 -1.006952
2 -0.03387729 -1.006952
.se.smooth.gam.s.Sepal.Width..lower .se.smooth.Species.lower
1 0.26156418 -1.263422
2 -0.05826436 -1.263422dfCompile.tree
> library(gam)
> gam.object <- gam::gam(Sepal.Length ~ gam::s(Sepal.Width) + Species,
data = iris)> dfCompile(tree.object) %>% length # needs package dplyr for "%>%"
[1] 4
> dfCompile(tree.object)$segments %>% head # needs package dplyr for "%>%"
.x .y .xend .yend .n
2 2.250 39.49191 2.250 102.16833 73
3 1.500 33.64903 1.500 39.49191 53
4 1.000 31.29592 1.000 33.64903 20
5 2.000 31.29592 2.000 33.64903 33
6 3.000 33.64903 3.000 39.49191 20
7 6.125 39.49191 6.125 102.16833 77
> dfCompile(tree.object)$labels %>% head # needs package dplyr for "%>%"
.x .y .label
1 4.1875 102.16833 Petal.Length<4.25
2 2.2500 39.49191 Petal.Length<3.4
3 1.5000 33.64903 Sepal.Width<3.25
4 6.1250 39.49191 Petal.Length<6.05
5 5.2500 27.04709 Petal.Length<5.15
6 4.5000 24.00201 Sepal.Width<3.05
> dfCompile(tree.object)$leaf_labels %>% head # needs package dplyr for "%>%"
.x .y .label .yval
4 1 31.29592 4.74 4.735000
5 2 31.29592 5.17 5.169697
6 3 33.64903 5.64 5.640000
10 4 22.26715 6.05 6.054545
11 5 22.26715 6.53 6.530000
12 6 24.00201 6.60 6.604000dfCompile.matrix
This function has an option type that takes either image or heatmap (see reference documentation).
Note that both also provide the potential for row and column associated dendrograms.
> dfCompile(iris_c, type = "heatmap")$df %>% head #needs package dplyr for "%>%"
.var1 .var2 value
1 Sepal.Width Sepal.Width 1.0000000
2 Sepal.Length Sepal.Width -0.1175698
3 Petal.Length Sepal.Width -0.4284401
4 Petal.Width Sepal.Width -0.3661259
5 Sepal.Width Sepal.Length -0.1175698
6 Sepal.Length Sepal.Length 1.0000000
> dfCompile(iris_c, type = "image")$df %>% head
.var1 .var2 value
1 Sepal.Length Sepal.Length 1.0000000
2 Sepal.Width Sepal.Length -0.1175698
3 Petal.Length Sepal.Length 0.8717538
4 Petal.Width Sepal.Length 0.8179411
5 Sepal.Length Sepal.Width -0.1175698
6 Sepal.Width Sepal.Width 1.0000000TODO:
Overarching (when making new object functionality):
- 1. check what
broomdoes for each object. Document whenbroomdoesn't create what we need for the visualizations.
ggDiagnose (models):
- 1.
lm,glm(fromstatspackage):ggDiagnose.lm - 2.
Gam(from the originalgampackage - notmgcv- or at least not first round) - 3.
glmnet(fromglmnetpackages):ggDiagnose.glmnet,ggDiagnose.cv.glmnet-
plot.mrelnet,plot.multnet needed?
-
- 4.
tree(fromtreepackage - could also one fromrpartpackage - useggdendroand see examples fromggdendro. - 5.
randomForest - 6.
mclust(multiple objects - multiple options forMclustgraphic. - will take time) - 7. PCA/factor analysis, etc objects
CM 36-402 models' from Cosma's book
- 8.
npregfromnp, kernel regression (pg 124)- looks like a 3d option (
plotly?) and a partial-dependence like plot
- looks like a 3d option (
- 9. tree stuff - plot slices 2d? (pg ~300)
ggVis (other objects):
- 1.
sp - 2.
dendrogram(very similar to tree, and there is already aggdendro... package that would do it better. Let's not do this for now) - 3.
matrix(forheatmap? orimage?) - let's do both for now.
packages that I'm not sure can be done:
- 1.
lme4(these objects don't seem to haveplot._functions to emulate.) - 2.
kde2d- from MASS (actually just produces a list ...) - butzcould be dealt with a matrix that you want to do "image" to. - 9.
ranger(better RF package?) (doesn't actually haveplotfunction for ranger objects...)
teaching:
- 1. In examples for each function provide code to create some / all of the plots in a more basic manner with straight use of
tidyverse. (Is this worthwhile? maybe for non-trivial/non standardggplot2graphics?)
best coding practices:
- 1. decide which parameters are passed to the visualization functions and how they differ / are the same of the
plotimplementation. For all visuals we haveshow.plotandreturnparameters.
documentation:
- 1. create a new file for package documentation. See: http://r-pkgs.had.co.nz/man.html#man-packages
things to look into:
S3 methods? - should the whole thing be a S3 method?
Write a blog post about putting a changing axis on top, link to the one where they transform the equation. Mention Hadley's thoughts on the matter.
ideas for tests:
- for all check the data frame coming out of
ggDiagnosevsdfCompile, - look at the names of the data frame returned by
dfCompile - check return options for
ggDiagnose
readme needs:
Probably should be showcasing dfCompile as well... how to do so in .md...