Working with multi-proxy paleo-archive data is inherently difficult. There are multiple locations, multiple parameters, and a lot of discipline-specific norms for plot layout and notation. This package simplifies a few workflows to promote the use of R and reproducible documents in paleo-based studies.
You can install tidypaleo from github with:
# install.packages("devtools")
devtools::install_github("paleolimbot/tidypaleo")To get the most out of strat diagrams, you will also need the patchwork package by [@thomasp85]((https://github.com/thomasp85). You can also install this from GitHub:
# install.packages("devtools")
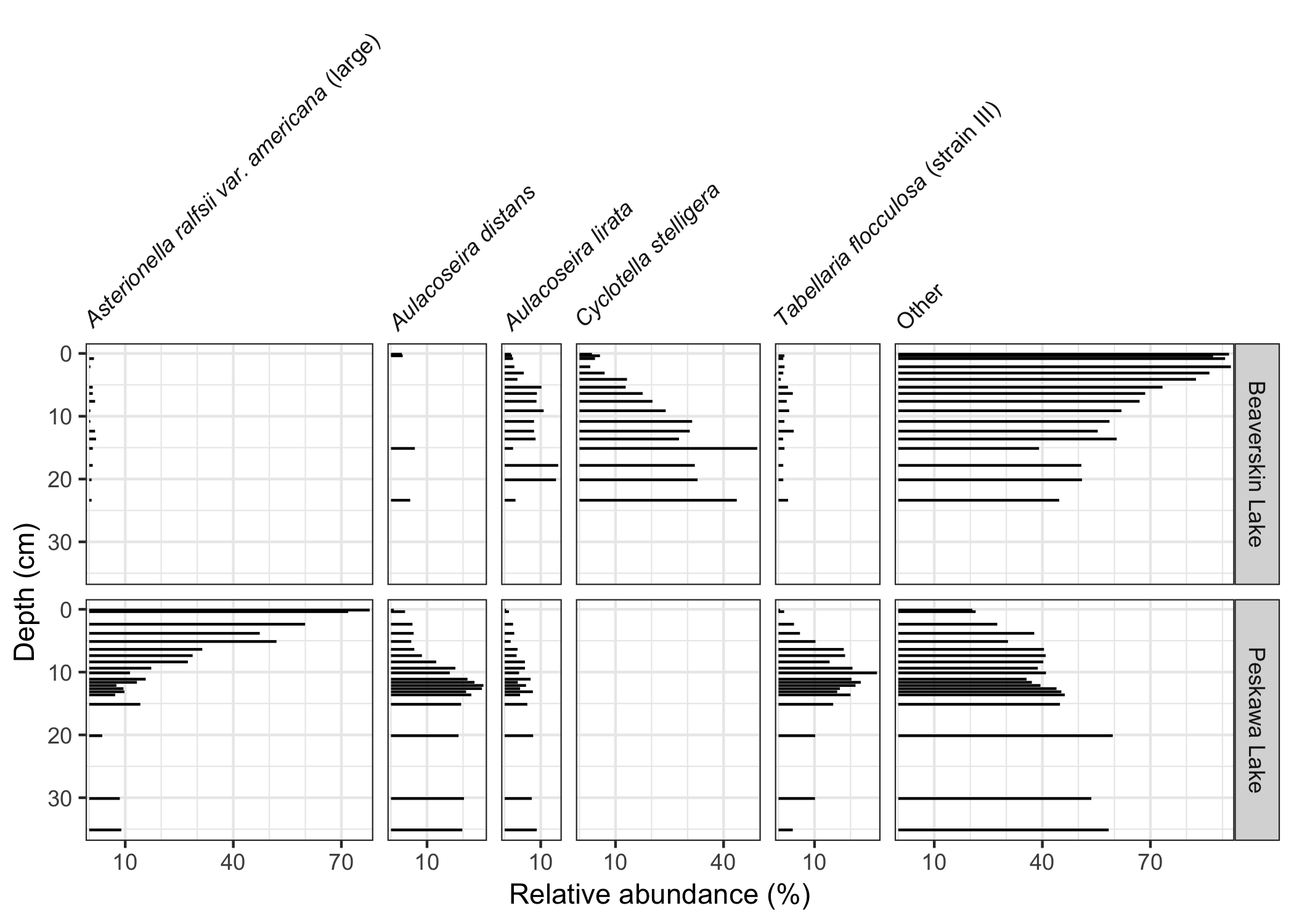
devtools::install_github("thomasp85/patchwork")This package exposes a number of functions useful when creating stratigraphic diatrams, including facet_abundanceh(), which combines several other functions to help create stratigraphic plots using ggplot2. The geom_col_segsh() geometry draws horizontal sements, which are commonly used to show species abundance data.
library(ggplot2)
library(tidypaleo)
theme_set(theme_bw())
ggplot(keji_lakes_plottable, aes(x = rel_abund, y = depth)) +
geom_col_segsh() +
scale_y_reverse() +
facet_abundanceh(vars(taxon), grouping = vars(location)) +
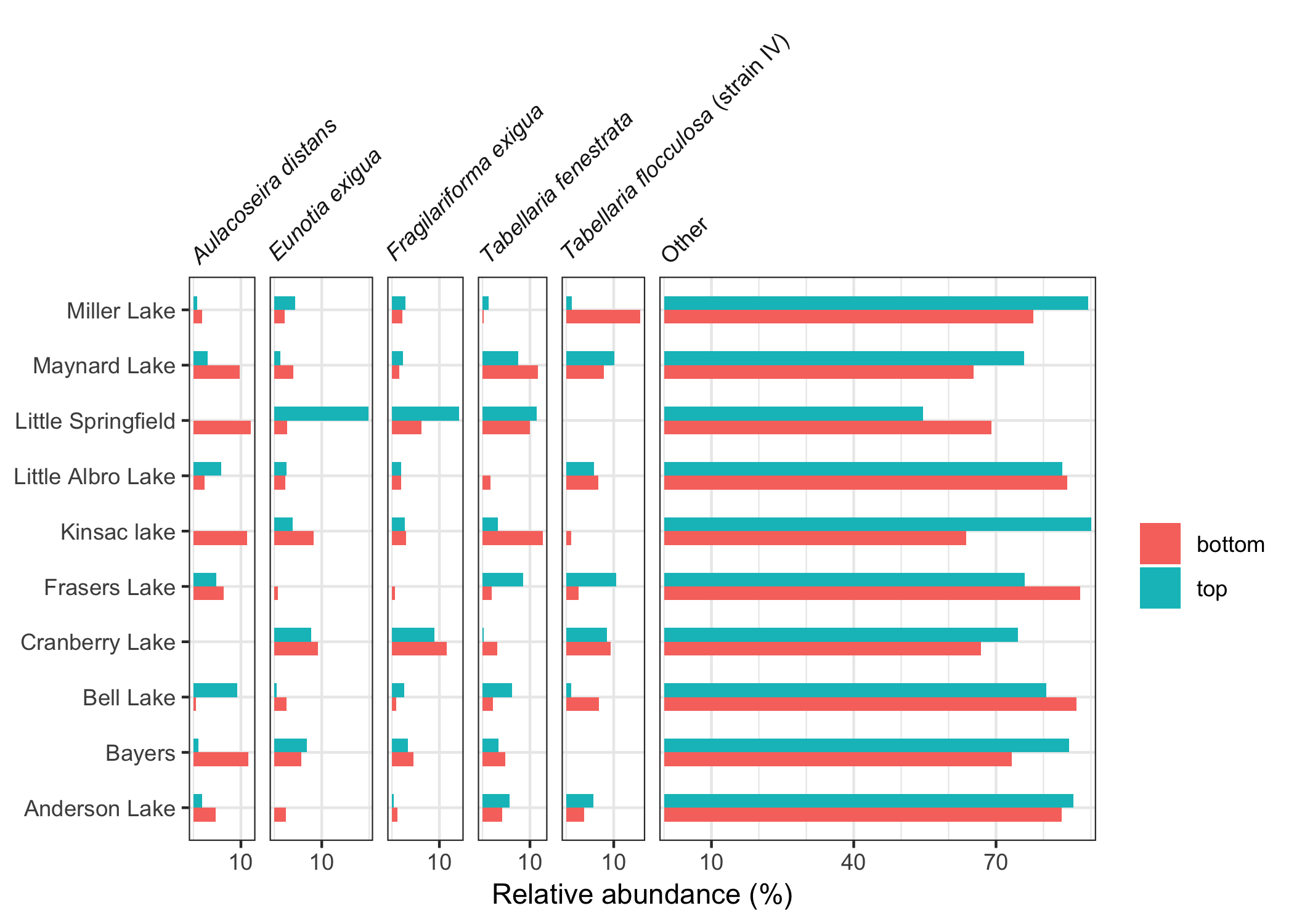
labs(y = "Depth (cm)")Relative abundance data with a discrete y axis can be drawn using geom_colh() and position_dodgev() from the ggstance package.
ggplot(halifax_lakes_plottable, aes(x = rel_abund, y = location, fill = sample_type)) +
geom_colh(width = 0.5, position = "dodgev") +
facet_abundanceh(vars(taxon)) +
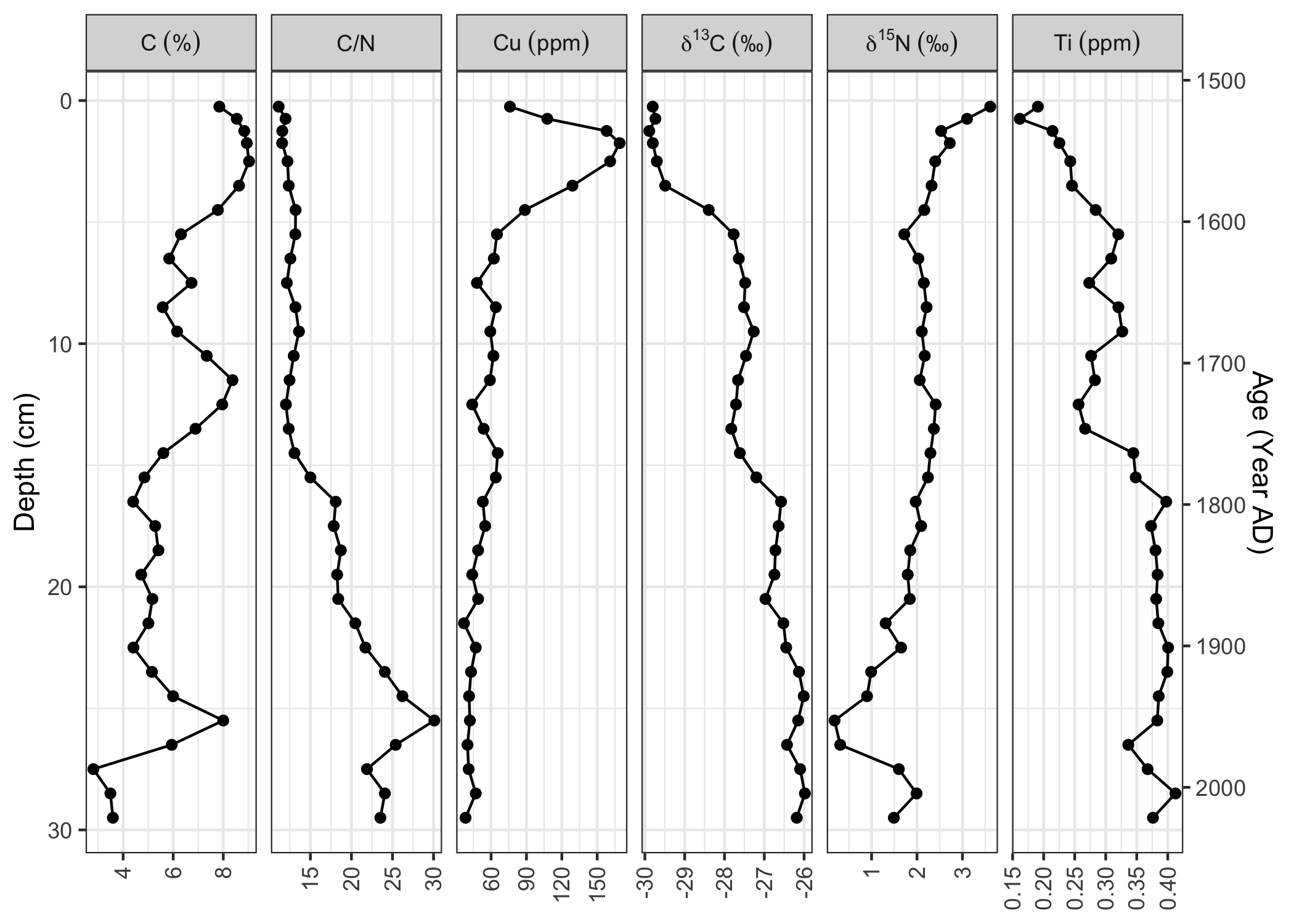
labs(fill = NULL, y = NULL)Age depth models can be constructed (using age_depth_model()) and used to draw second axes on stratigraphic plots (using scale_y_depth_age()). The facet_geochem() function wraps facet_wrap(), and includes a labeller (label_geochem) that can add units, and automatically renames d15N, d13C and d18O such that they are labelled correctly.
alta_adm <- age_depth_model(
alta_lake_bacon_ages,
depth = depth_cm,
age = 1950 - age_weighted_mean_year_BP
)
ggplot(alta_lake_geochem, aes(x = value, y = depth)) +
geom_lineh() +
geom_point() +
scale_y_depth_age(alta_adm, age_name = "Age (Year AD)") +
facet_geochem_wraph(
vars(param),
nrow = 1,
units = c(
C = "%",
Cu = "ppm",
Ti = "ppm",
d15N = "‰",
d13C = "‰"
)
) +
labs(y = "Depth (cm)", x = NULL)