The Non-coding RNA Databases Resource (NRDR) is a source of data related to over 100 non-coding RNA (ncRNA) databases available over the internet. Our ncRNA Databases Resource tries to represent an updated source for databases information retrieval. It is part of a review with the aim of surveying all ncRNA databases available.
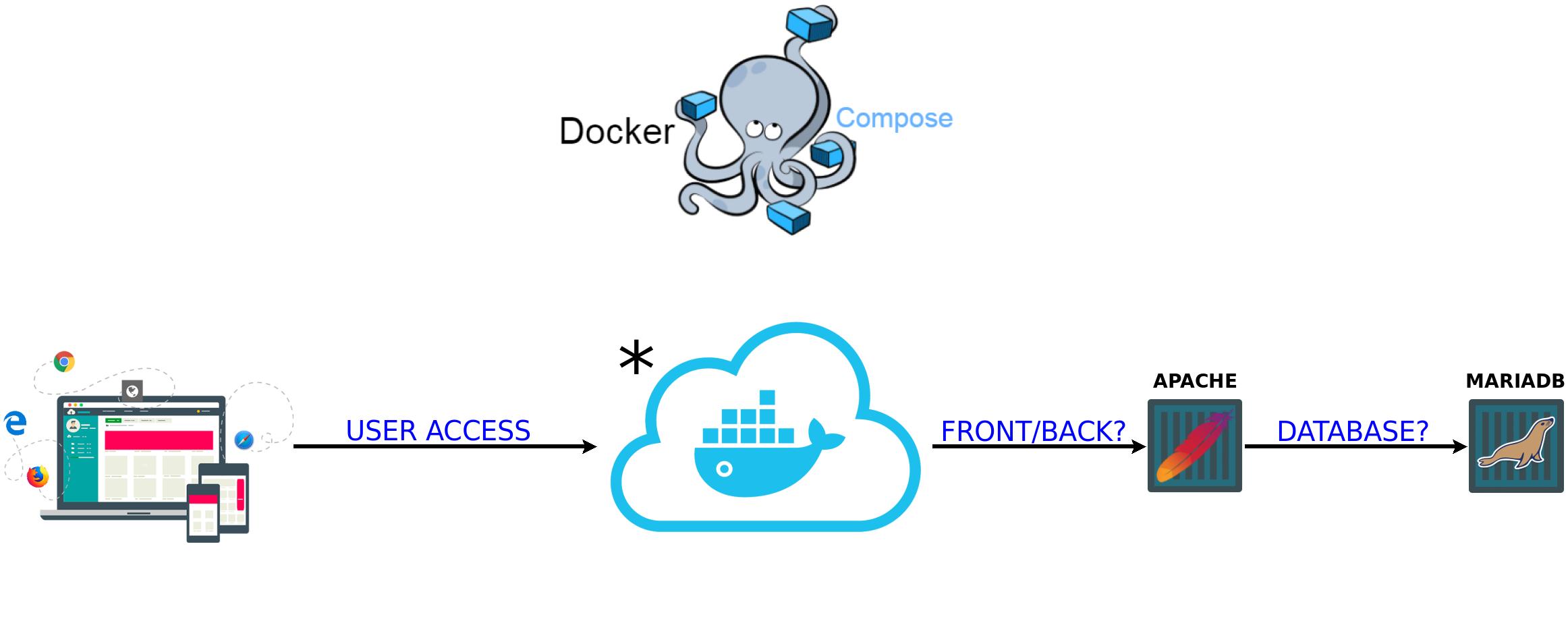
This cloud represents the proxy environment and maintenance of SSL certificates that handles requests and forwards them to the project. This environment is present in the following repository: Reverse Provy Apps .
- Frontend - Apache + PHP;
- Database - MariaDB.
2 Containers.
- Git version 2 or above
- Docker version 17.09.1-ce or above (https://docs.docker.com/install/)
- Docker Compose 1.20.1 or above (https://docs.docker.com/compose/install)
Clone the repository with:
user@host:~# git clone https://gitlab.com/integrativebioinformatics/nrdr.git
Create file .env in root directory on repository informing enviremont variables, content example:
user@host:~/nrdr# cat .env
DB_NAME=database_name
DB_USER=database_user
DB_PASS=database_passwordCopy content frontend files PsalmonisDB for directory volumes/frontend/, thus:
user@host:~/nrdr# ls -l volumes/frontend/
total 56
drwxr-xr-x. 14 amandabackup 33 4096 Abr 18 00:17 application
drwxrwxr-x. 7 amandabackup 33 4096 Abr 17 20:47 assets
-rw-r--r--. 1 amandabackup 33 557 Abr 18 00:17 composer.json
-rw-r--r--. 1 amandabackup 33 10333 Abr 18 00:17 index.php
-rw-r--r--. 1 amandabackup 33 13319 Abr 18 00:17 license.txt
drwxr-xr-x. 2 amandabackup 33 4096 Abr 25 12:36 sessions
drwxr-xr-x. 8 amandabackup 33 4096 Abr 18 00:17 system
drwxr-xr-x. 3 amandabackup 33 4096 Abr 18 00:17 webservicesDefine permissions for user www-data in directory back/front which will be mounted as volume in container. Because the user may not exist on the host host, we use the gid that is standard on any system. Execute:
user@host:~/nrdr# chown 33:33 -R volumes/frontendDatabase files?
Just add their sql files in the build/db/dump/ directory in deploy case, they will be executed in the container in alphabetical order. So if you have a schema.sql and data.sql file, is important renaming them to 1-schema.sql and 2-data.sql.
If you want, you can compress them (to reduce space) in a format accepted by the docker (gzip, bzip2 our xz) and the container will do the extraction.
OBS.: The content on /var/lib/mysql in "real time" container, will be in volumes/db .
In the root repository, execute the next command:
user@host:~/nrdr# docker-compose up --build -dThe option -d execute containers in background.
Is possible check process with:
user@host:~/nrdr# docker-compose logs -fThe option -f stay logs in screen, to cancel execute Ctrl + C.
OBS.: For scalability reasons, this project was designed to receive requests from a proxy, so port 80 is not exposed. If you need to run the environment locally (exposed port 80), the command is as follows:
user@host:~/nrdr# docker-compose -f docker-compose-local.yml up -d