Codebase accompanying the NeurIPS 2024 paper "Distributed Memory through the Lens of Random Features"
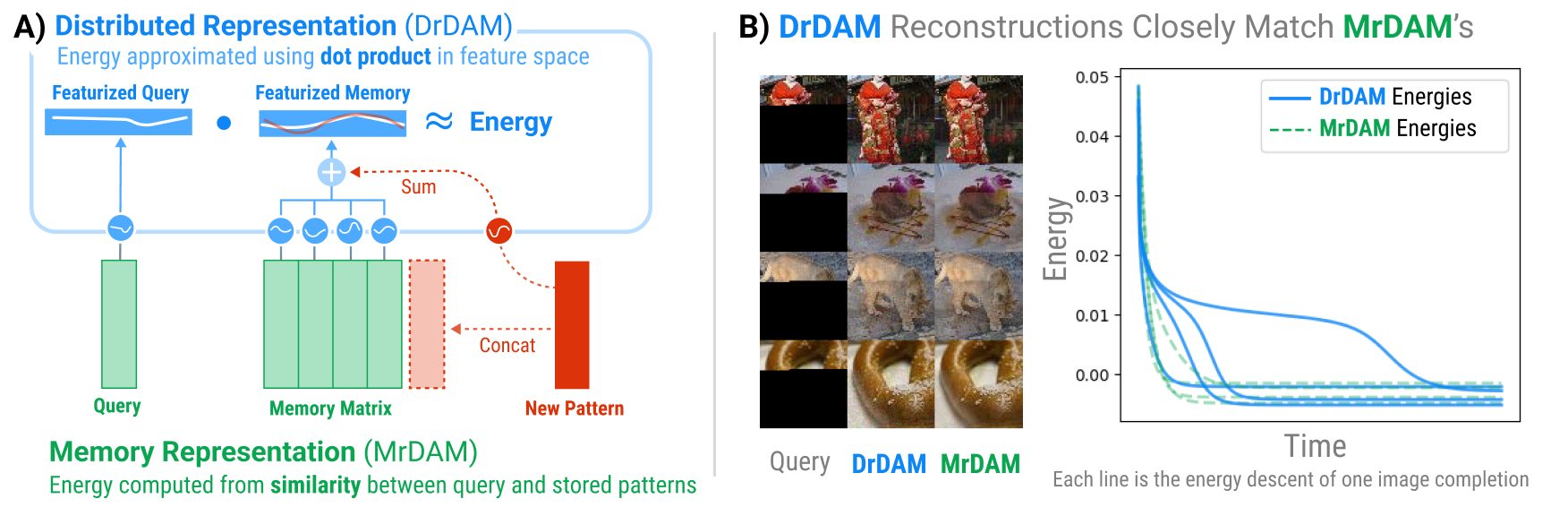
DrDAM is the first technique to show how memories can be distributed in Dense Associative Memories (DenseAMs), much like how memories were distributed in the original Hopfield network. The traditional Memory Representation for Dense Associative Memory (MrDAM) is a "slot-based" associative memory where each memory is represented as a "slot" (i.e., row or column) of a weight matrix. New memories are added by concatenating a new vector to an existing weight matrix. DrDAM takes advantage of Random Features to store patterns via summation into a weight tensor of constant size rather than concatenation. See Figure 1 below (from the paper) for an illustration. DrDAM closely approximates both the energy and fixed-point dynamics of the traditional Memory Representation for Dense Associative Memory (MrDAM) while having a parameter space of constant size.
Figure 1: The Distributed Representation for Dense Associative Memory (DrDAM) approximates both the energy and fixed-point dynamics of the traditional Memory Representation for Dense Associative Memory (MrDAM) while having a parameter space of constant size.This repository contains the code for recreating all experiments of the main paper. Unless otherwise noted, all reported results were created on a single L40s GPU with ~48GB of VRAM.
conda env create -f environment.yml
conda activate distributed_DAM
pip install -r requirements_base.txt # For pip package, just to use DrDAM tooling
pip install -r requirements_additional.txt # For full experiments
pip install --upgrade "jax[cuda12]" # Match CUDA version to your GPU
make data # Download data. Takes ~10 min depending on internet speedIf you don't care about running the full experiments, you can install the package for just the tooling.
git clone https://github.com/bhoov/distributed_DAM.git
cd distributed_DAM
pip install -e .or
pip install git+https://github.com/bhoov/distributed_DAM.gitQuick Usage After installing,
from distributed_DAM import SIM_REGISTRY as KernelOpts
import jax.random as jr
drdam = KernelOpts['SinCosL2DAM'](jr.PRNGKey(0), 10, 100, beta=1.)We follow a "shallow" directory structure that obviates the need for submodules and editable pip installations. Run all commands from the root directory. Unless otherwise mentioned, experiments default to running on GPU0.
Recreate the trajectories shown in the right half of Fig 1 of our paper by running the following code:
python exp_PINF.py fig1
- Output figures saved in
figs/PINF
(used an A100 with 80GB of VRAM for this experiment)
python exp_PINF.py fig2
- Output figure saved in
figs/PINF2
(experiment takes several days as currently implemented) Data is first analyzed across a wide range of configurations. Note that m is our old notation for Y in the paper.
python exp_QUANT1b_opt_retrieval.py --betas 10 30 40 50 --ms 5000 40000 80000 120000 160000 200000 300000 400000 500000 --outdir results/QUANT1b_near_0.1_retrieval --do_retrieval
- Saves data to
results/QUANT1b_near_0.1_retrieval
Once the data is generated, we analyze it for Figs 3 and 4A.
python eval_QUANT1b_no_retrieval+bounds.py (Fig 3)
python eval_QUANT1b_retrieval.py (Fig 4A)
- Saves figures to
figs/QUANT1b
(experiment takes ~10 minutes)
python exp_QUAL1__qualitative_imgs.py
- Output figure saved in
figs/QUAL1
(experiment takes ~2 minutes)
python exp_QUANT1c.py
python eval_QUANT1c.py (Fig 5)
- Saves figures to
figs/QUANT1c
(experiment takes ~2 minutes)
python exp_QUANT2__kernel_ablations.py
python eval_QUANT2__kernel_ablations.py
- Runs on CPU by default
- Saves figures to
figs/QUANT2
@inproceedings{
hoover2024dense,
title={Dense Associative Memory Through the Lens of Random Features},
author={Benjamin Hoover and Duen Horng Chau and Hendrik Strobelt and Parikshit Ram and Dmitry Krotov},
booktitle={The Thirty-eighth Annual Conference on Neural Information Processing Systems},
year={2024},
}