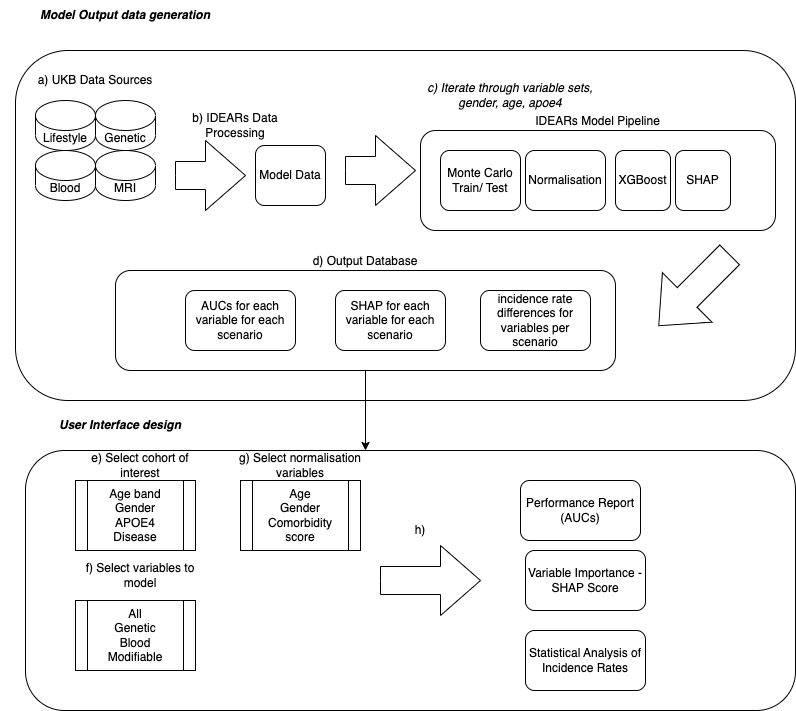
This is the codebase for IDEARs - The Integrated Disease Explanation and Associations Risk Scoring. Its overall architecture is shown below:
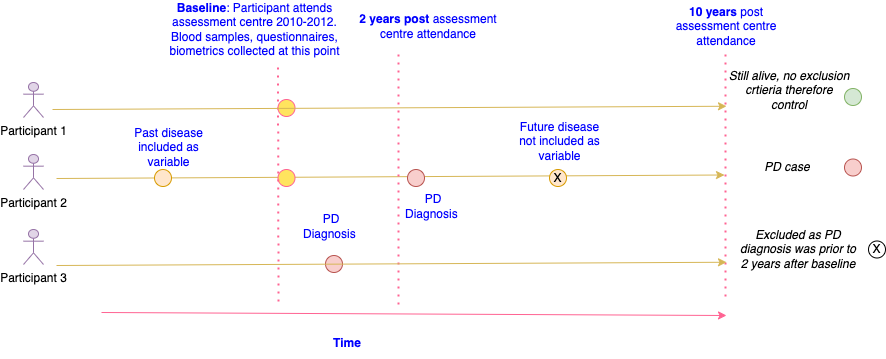
The code is designed to represent the following situation for prospective studies, which depicts a participant in UKB attending the centre at baseline and then subsequently having a number of outcomes occur
To ease the configuation, please install Anaconda and set this up in a virtual environment.
- Install Anaconda:
https://www.anaconda.com/products/individual
Import modules etc.
This folder shows the implementation of the IDEARs platform.
📦ukb_IDEARS-pipeline-poc
┣
┃ ┣ src
┃ ┃ ┣ idears
┃ ┃ ┃ ┣ 📂 preprocessing
┃ ┃ ┃ ┃ ┣ 📜 data_proc.py
┃ ┃ ┃ ┃ ┣ 📜 idears_backend.py
┃ ┃ ┃ ┃ 📂 models
┃ ┃ ┃ ┃ ┣ 📜 mlv2.py
┃ ┃ ┃ ┃ 📂 frontend
┃ ┃ ┃ ┣ ┣ 📜 app1.py
┃ ┣ applications
┃ ┃ ┃-AD
┃ ┃ ┃-PD
┣ 📜config.yaml
┣ 📜requirements.txt
┣ 📜main.py
┣ 📜README.md
┣
Note for Parkinson's please go to the following link to see the notebooks used to generate the data in our manuscript
"Machine Learning Analysis of the UK Biobank Reveals IGF-1 and Inflammatory Biomarkers Predict Parkinson’s Disease Risk"
https://github.com/MikeAllwright23/idears_orig/tree/main/notebooks/pd
The data behind the figures is also available at this location
https://github.com/MikeAllwright23/idears_orig/tree/main/data
Michael Allwright - michael@allwrightanalytics.com, michael.allwright@sydney.edu.au