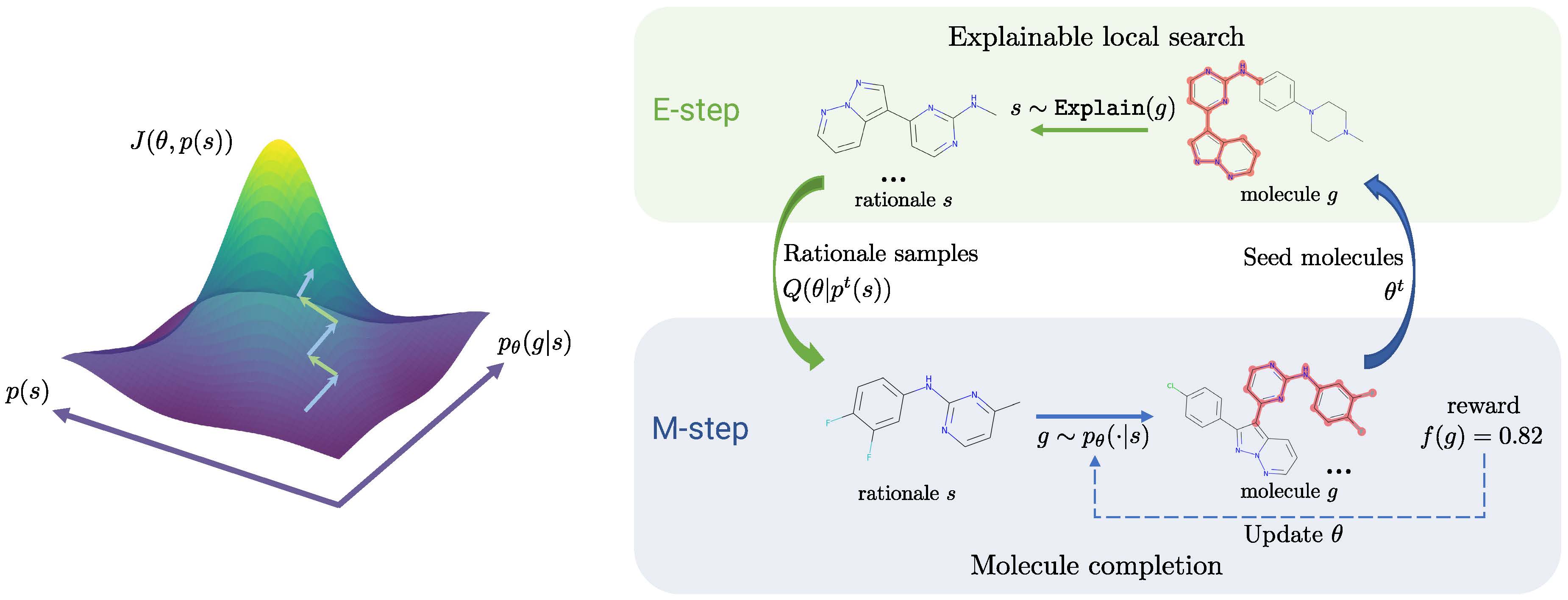
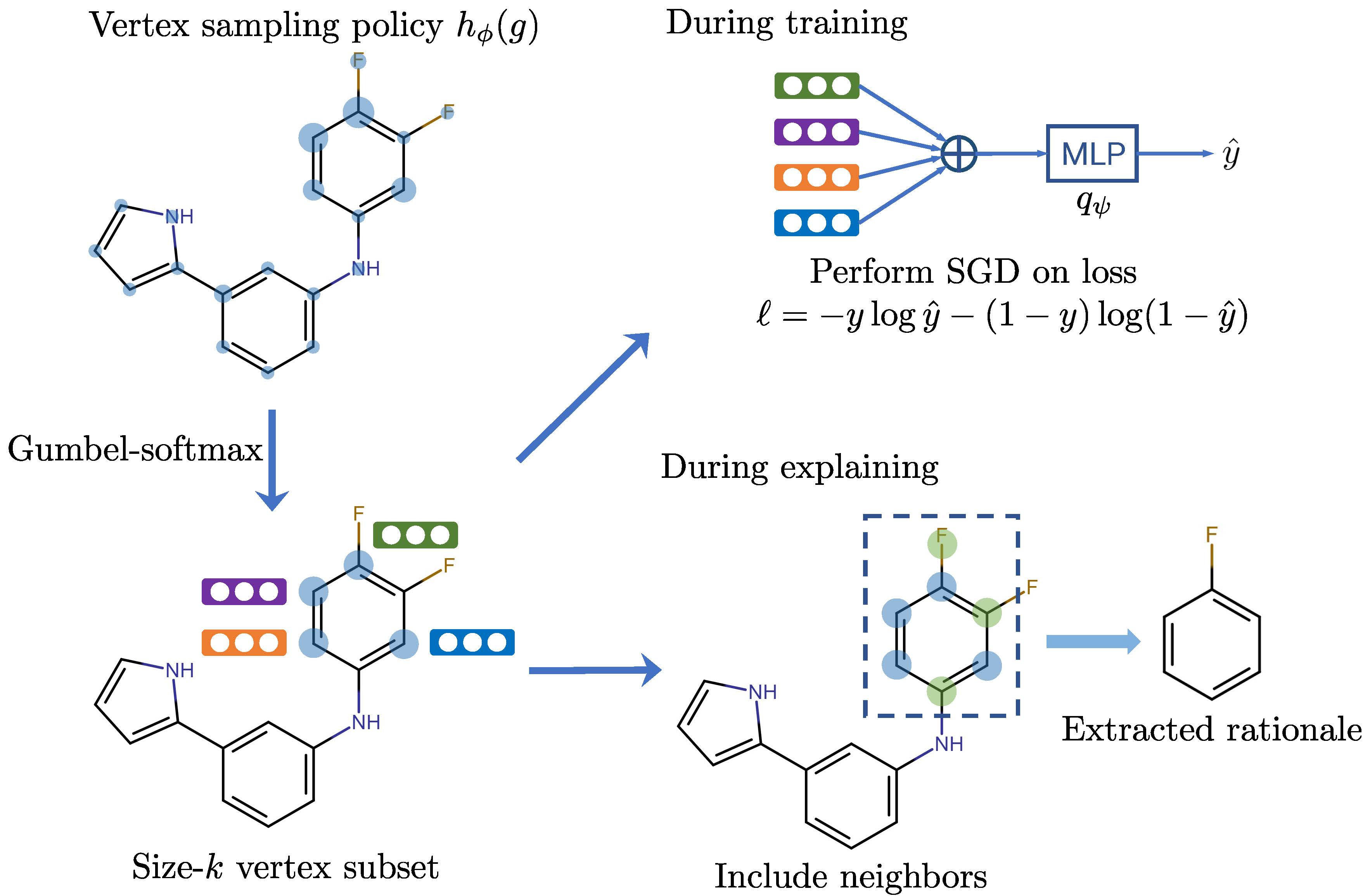
Molecule Optimization by Explainable Evolution
@inproceedings{Chen2021MolEvol,
title={Molecule Optimization by Explainable Evolution},
author={Binghong Chen* and Tianzhe Wang* and Chengtao Li and Hanjun Dai and Le Song},
booktitle={International Conference on Learning Representations},
year={2021}
}
Overview
We release the PyTorch code for the MolEvol. [Paper]:
Details for Graph Explanation Process
Install Dependencies
You should install dependencies for chemprop and multiobj-rationale first.
Install chemprop
conda create -n chemprop python=3.8
conda activate chemprop
conda install pytorch==1.6.0 torchvision==0.7.0 -c pytorch
conda install -c conda-forge rdkit
pip install -r requirements.txt
pip install chemprop
Install multiobj-rationale
cd ..
git clone git@github.com:anonymous20201002/multiobj-rationale.git
cd multiobj-rationale
pip install -e .
cd ../mol_finder
How to Use
For instance, if you want to jointly optimize jnk3, gsk3, QED, SA for the molecules.
Run the following command:
export CUDA_VISIBLE_DEVICES=0
export TF_CPP_MIN_LOG_LEVEL=2
export PYTHONPATH=$PWD
python mol_finder/main.py --rounds=10 --epoch=1
You will get the molecule optimization model along with the optimized molecules in the experiment folder.