omplotr: 'ggplot2' Based RNAseq Plot Function Collection
Installation and loading
Install the latest version from GitHub as follow:
# Install
if(!require(devtools)) install.packages("devtools")
devtools::install_github("bioShaun/omplotr")Theme
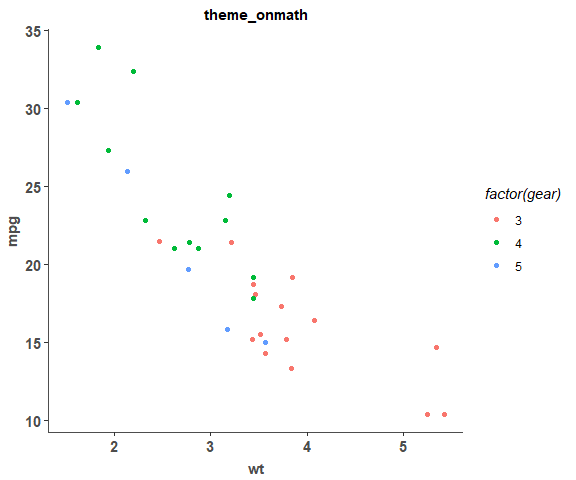
theme_onmath is a ggplot theme used in almost all rnaseq plots.
library(omplotr)
p <- ggplot(mtcars) + geom_point(aes(x = wt, y = mpg,colour = factor(gear)))
p + theme_onmath() + ggtitle("theme_onmath")Plot
functions to generate plot in ngs analysis
QC
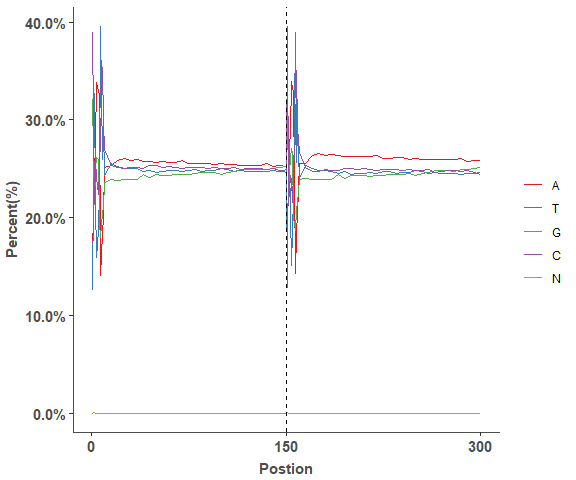
Reads GC distribution
# Fastqc GC result
head(gc_test_data, 4)
#> X.Base sample variable value
#> 1 1 DZ-A-LDY-1 A 0.1644290
#> 2 2 DZ-A-LDY-1 A 0.2451466
#> 3 3 DZ-A-LDY-1 A 0.2764013
#> 4 4 DZ-A-LDY-1 A 0.3381226
# lineplot of GC distribution across Fastq file
gc_line_plot(gc_test_data)Reads Quality barplot
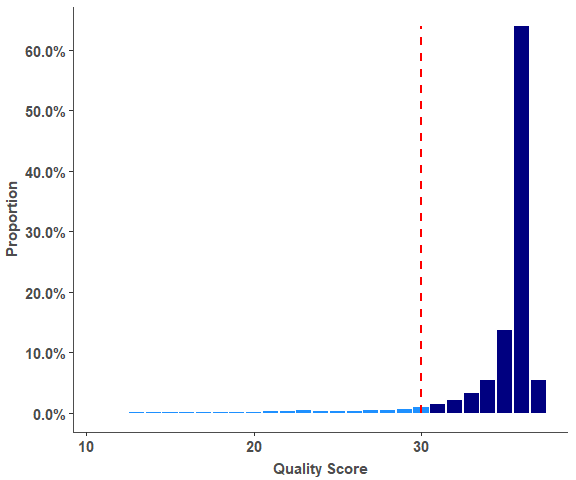
# Reads Quality result
# Bars of Quality <= 30 were marked with color 'dodgerblue',
# Bars of Quality > 30 were marked with color 'navy'.
head(rq_test_data, 4)
#> Quality Count Proportion color sample
#> 1 11 57 1.016564e-06 dodgerblue YP-B-WYX-6
#> 2 12 7352 1.311189e-04 dodgerblue YP-B-WYX-6
#> 3 13 40981 7.308736e-04 dodgerblue YP-B-WYX-6
#> 4 14 57256 1.021129e-03 dodgerblue YP-B-WYX-6
# Reads Quality barplot
reads_quality_plot(rq_test_data)Quant
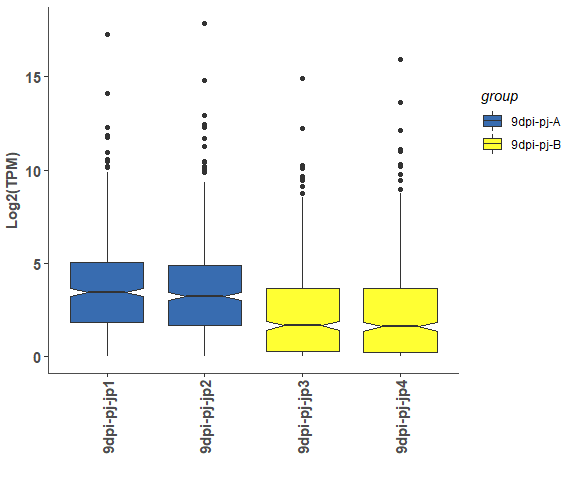
expression box, violin and density plot
# expression matrix
head(exp_test_data, 4)
#> 9dpi-pj-jp1 9dpi-pj-jp2 9dpi-pj-jp3 9dpi-pj-jp4
#> ENSRNA049464904 64.515 48.860 34.595 25.636
#> ENSRNA049468231 17763.048 28554.280 4878.607 12802.249
#> ENSRNA049468277 544.106 1152.839 169.713 497.665
#> ENSRNA049471043 4926.117 7815.150 1198.127 4545.421
# sample information
head(test_sample_data, 4)
#> condition sample
#> 1 9dpi-pj-A 9dpi-pj-jp1
#> 2 9dpi-pj-A 9dpi-pj-jp2
#> 3 9dpi-pj-B 9dpi-pj-jp3
#> 4 9dpi-pj-B 9dpi-pj-jp4
# boxplot
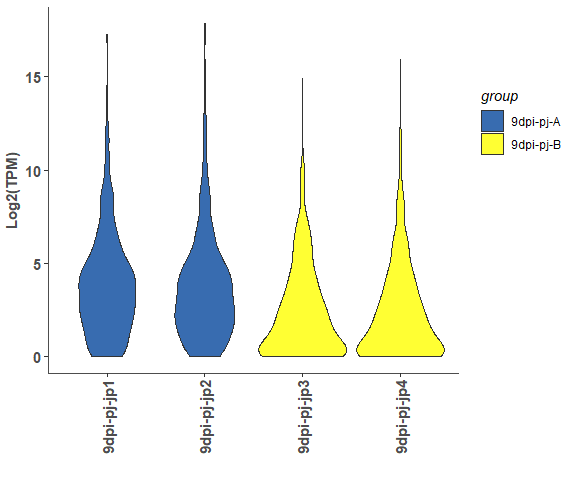
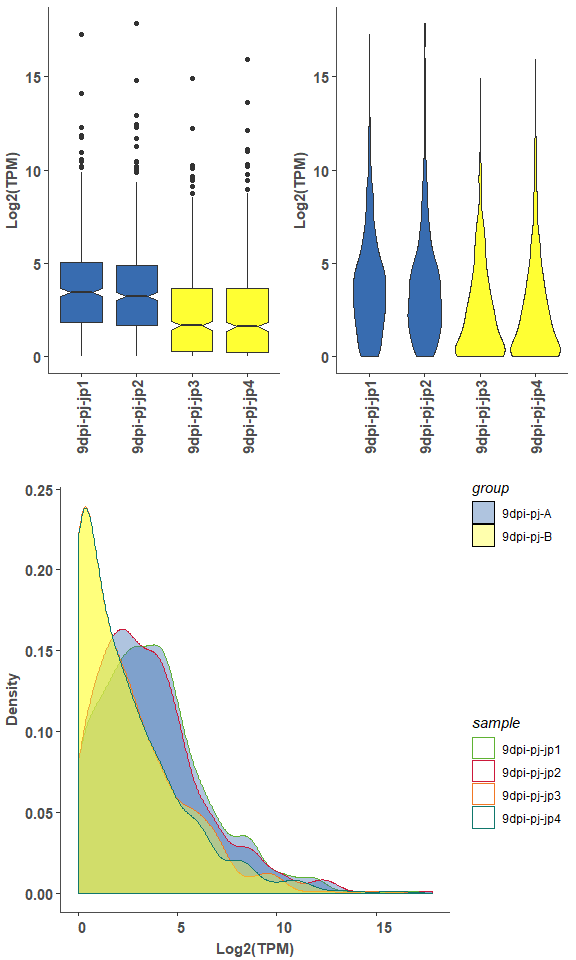
om_boxplot(exp_test_data, test_sample_data, 'box')# violin
om_boxplot(exp_test_data, test_sample_data, 'violin')# density
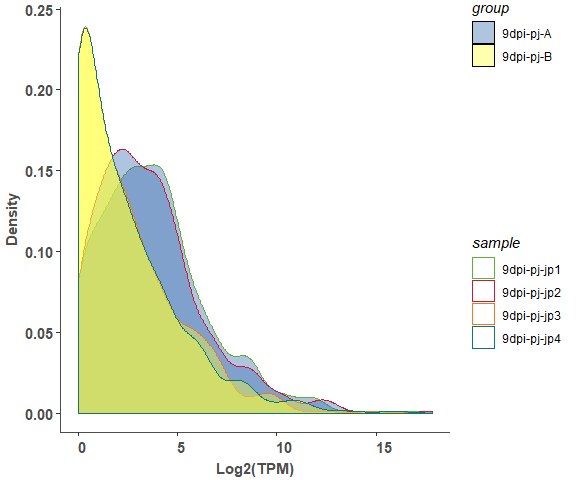
om_boxplot(exp_test_data, test_sample_data, 'density')# merged plot
om_boxplot(exp_test_data, test_sample_data, 'all')expression PCA analysis point plot
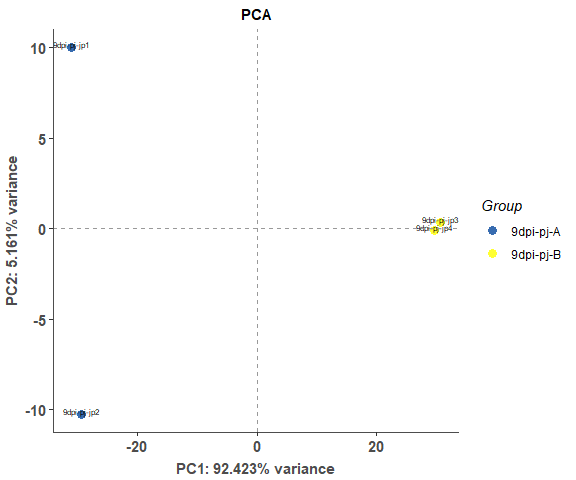
om_pca_plot(exp_test_data, test_sample_data)expression correlation heatmap
om_correlation_plot(exp_test_data, test_sample_data)diff expression volcano plot
# diff result
head(diff_test_data, 4)
#> Gene_ID X9dpi.pj.jp1 X9dpi.pj.jp2 X9dpi.pj.jp3
#> 21437 Os01g0977250 4.780 4.551 7.806
#> 33806 Os03g0738600 1.209 0.946 1.338
#> 30663 Os03g0823900 5.598 5.303 6.322
#> 32700 EPlOSAG00000051674 0.000 0.000 1.482
#> X9dpi.pj.jp4 logFC PValue FDR compare
#> 21437 6.489 -0.21257899 0.3964249 0.6631611 Case1_vs_Control
#> 33806 1.392 0.03988567 0.9082303 1.0000000 Case1_vs_Control
#> 30663 7.596 0.04831892 0.8428859 0.9858939 Case1_vs_Control
#> 32700 0.000 -2.62518576 1.0000000 1.0000000 Case1_vs_Control
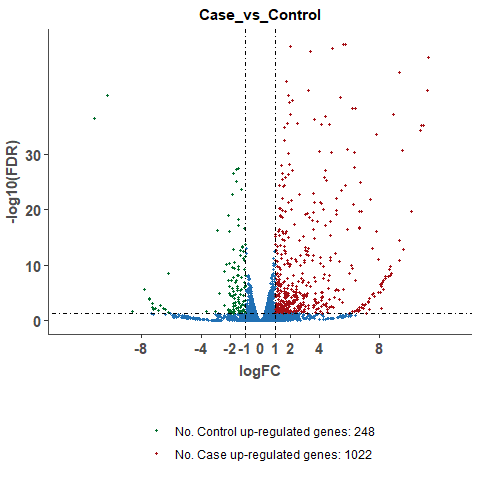
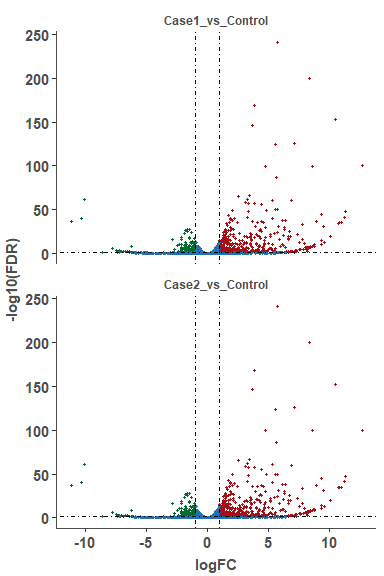
# plot volcano plot for a single compare
om_volcano_plot(diff_test_data, 'Case_vs_Control')# plot volcano plot for merged results
om_volcano_plot(diff_test_data, 'ALL')expression heatmap
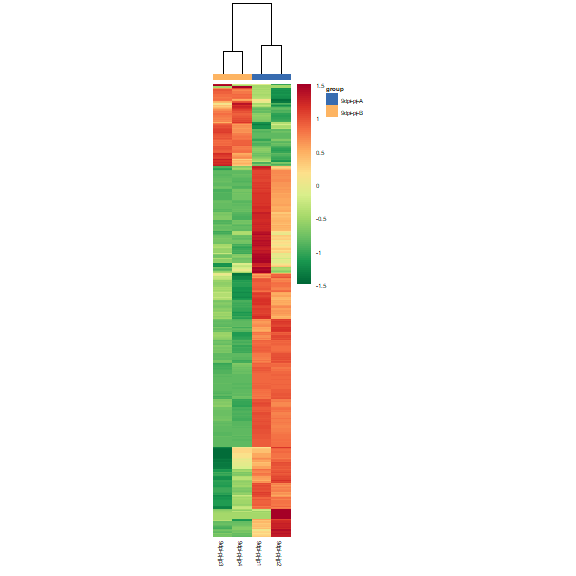
# plot expression heatmap
om_heatmap(exp_test_data, test_sample_data)expression cluster line plot
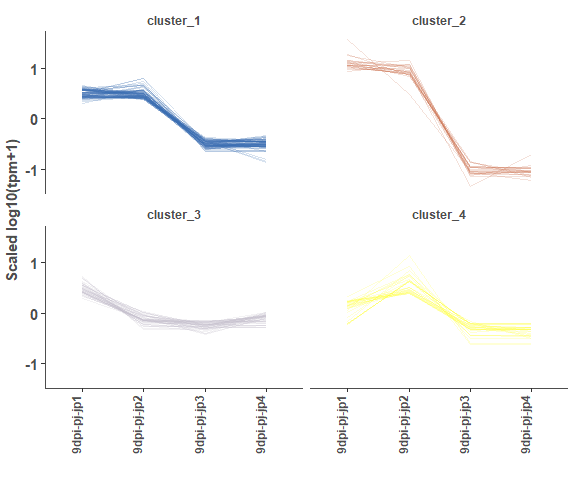
# cluster result
head(cluster_test_data, 4)
#> cluster Gene_id variable value
#> 1 cluster_1 EPlOSAG00000008604 9dpi-pj-jp1 0.4935853
#> 2 cluster_1 EPlOSAG00000018483 9dpi-pj-jp1 0.5236810
#> 3 cluster_1 EPlOSAG00000027962 9dpi-pj-jp1 0.5008437
#> 4 cluster_1 EPlOSAG00000045132 9dpi-pj-jp1 0.4879498
# cluster plot
om_cluster_plot(cluster_test_data)Enrichment analysis
GO enrichment analysis
# diff genes
test_diff_genes <- go_test_data_list[['test_diff_genes']]
head(test_diff_genes, 4)
#> [1] "Os01g0150000" "Os01g0172600" "Os01g0220700" "Os01g0303600"
# gene length
test_gene_len <- go_test_data_list[['test_gene_len']]
head(test_gene_len, 4)
#> gene_id gene_len
#> 1 Os01g0290700 2289
#> 2 Os01g0249200 5219
#> 3 Os01g0152200 3747
#> 4 Os01g0295700 5836
# get go annotation file
gene_go_map <- system.file("extdata", "topgo_test_data.txt", package = "omplotr")
gene_go_map_df <- data.table::fread(gene_go_map, header = F)
head(gene_go_map_df, 4)
#> V1 V2
#> 1: Os01g0166300 GO:0017176
#> 2: Os01g0296700 GO:0016787,GO:0005975,GO:0004553
#> 3: Os01g0290700 GO:0005524,GO:0016887
#> 4: Os01g0236400 GO:0005737,GO:0005856
# run goseq and show result
goseq_output <- om_goseq(test_diff_genes, test_gene_len, gene_go_map)
head(goseq_output, 4)
#> category over_represented_pvalue qvalue numDEInCat numInCat
#> 571 GO:0010287 0.008881002 0.3812842 2 2
#> 848 GO:0042803 0.008933210 0.3812842 2 2
#> 76 GO:0001172 0.024128606 0.3812842 2 3
#> 123 GO:0003968 0.024128606 0.3812842 2 3
#> term ontology
#> 571 plastoglobule CC
#> 848 protein homodimerization activity MF
#> 76 transcription, RNA-templated BP
#> 123 RNA-directed RNA polymerase activity MF
#> DE_id
#> 571 Os01g0173000,Os01g0118000
#> 848 Os01g0235200,Os01g0251000
#> 76 Os01g0198000,Os01g0198100
#> 123 Os01g0198000,Os01g0198100
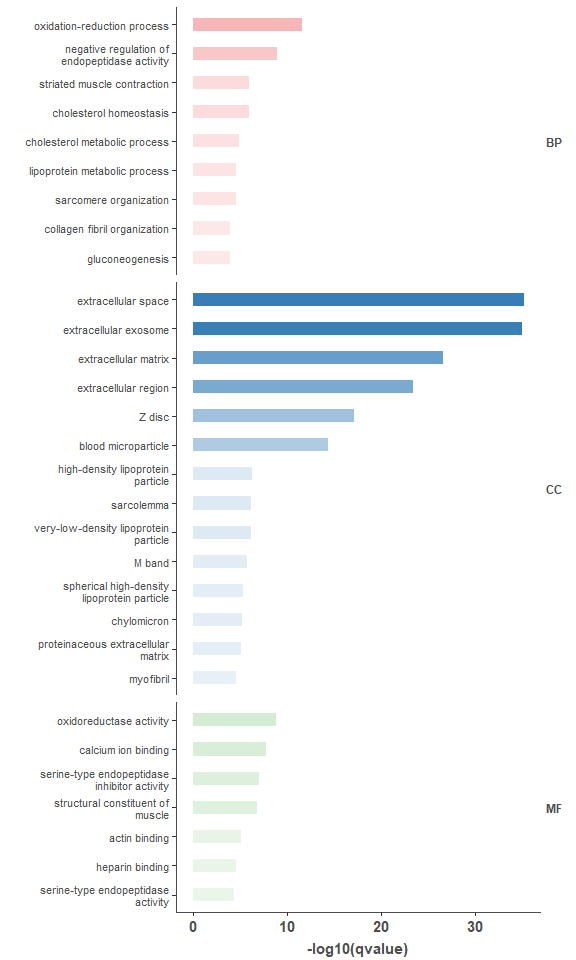
# go enrichment bar plot
go_enrich_file <- system.file("extdata", "enrichment",
"example.go.enrichment.txt",
package = "omplotr")
go_enrich_list <- clean_enrich_table(go_enrich_file)
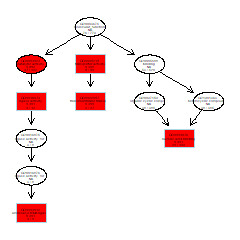
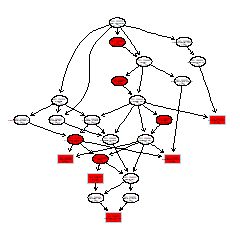
om_enrich_bar_plot(go_enrich_list$table, ylab_title=go_enrich_list$title)GO DAG graph
# run topGO
om_topgo(gene_go_map, test_diff_genes, goseq_output)Mapping
mapping_stats <- system.file("extdata", "all_sample.mapping.xls", package = "omplotr")
# show mapping stats table
mapping_df <- read.delim(mapping_stats)
head(mapping_df, 4)
#> Item A F1 K1 M1 M
#> 1 sequences: 95851866 102408480 88220198 102921732 89688164
#> 2 reads mapped: 94204523 98758455 87279503 101428039 88480886
#> 3 reads mapped and paired: 93189512 98569864 87122868 101234396 87992732
#> 4 reads unmapped: 1647343 3650025 940695 1493693 1207278
#> W
#> 1 89827664
#> 2 88521129
#> 3 88075758
#> 4 1306535
# show mapping summary
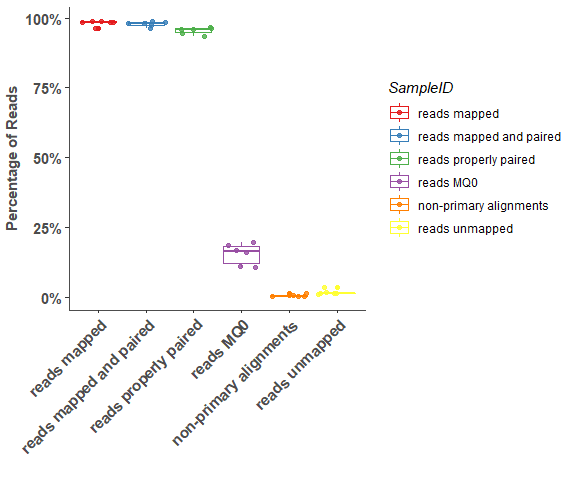
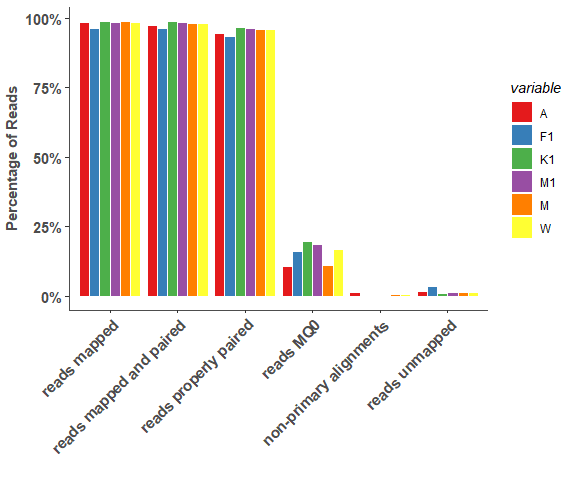
om_bwa_mapping_plot(mapping_stats, 5)# show detail sample information
om_bwa_mapping_plot(mapping_stats, 10)Reads Coverage
cov_stats <- system.file("extdata", "all_sample.genome.cov.xls", package = "omplotr")
# show coverage table
cov_df <- read.delim(cov_stats)
head(cov_df, 4)
#> Depth A F1 K1 M1 M W
#> 1 1 876435 764784 944681 786463 814983 857249
#> 2 2 630273 573112 761490 564038 600328 635532
#> 3 3 553156 479804 677398 484714 513316 544938
#> 4 4 523375 436214 652812 453986 470339 503152
# show coverage summary
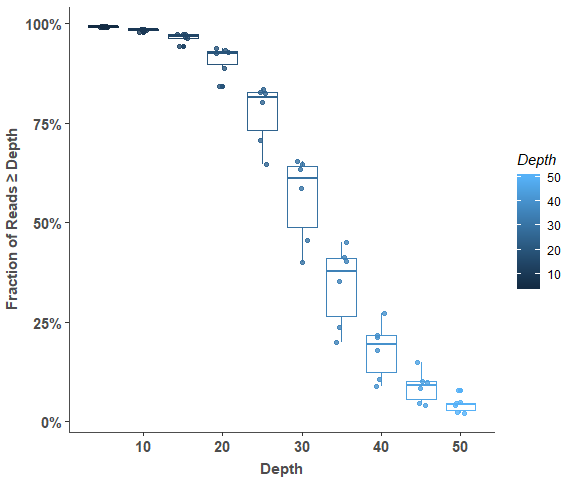
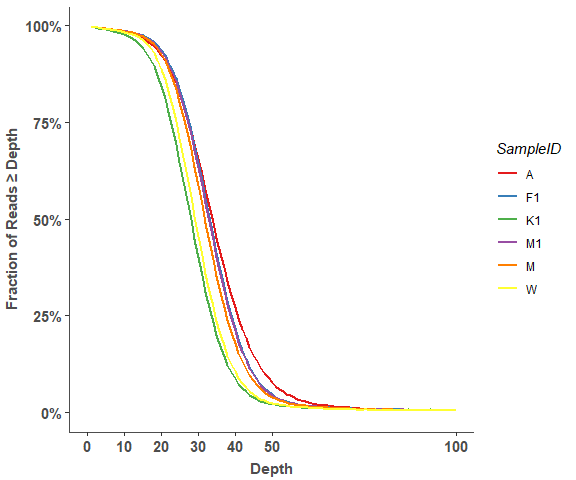
om_reads_cov_plot(cov_stats, 5, 100)# show detail sample information
om_reads_cov_plot(cov_stats, 10, 100)Variant Distribution
# transform variant summary data
stats_names <- om_const_reseq_variant[['var_file_labs']][1:2]
stats_dir <- system.file("extdata", "variant_stats",
package = "omplotr")
impact_map_file <- system.file("extdata", "variant_stats", "snpeff_varEffects.csv",
package = "omplotr")
test_pie_stats <- lapply(stats_names, om_var_pie_stats,
stats_dir=stats_dir,
impact_map_file=impact_map_file)
head(test_pie_stats[[1]], 4)
#> Type variable value Percentage fig
#> 1 DEL A 0.09495793 9.50% varType
#> 2 INS A 0.09794564 9.79% varType
#> 3 SNP A 0.80709642 80.71% varType
#> 4 DEL F1 0.09068346 9.07% varType
# plot pie plot for each sample
a_om_test_var_stats <- dplyr::filter(om_test_var_stats[[1]], variable == 'A')
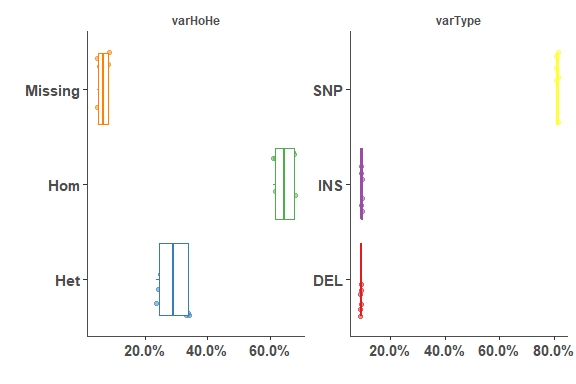
om_var_pie_plot(a_om_test_var_stats)# variant summary boxplot
om_test_var_stats_df <- plyr::ldply(om_test_var_stats, data.frame)
om_var_summary_plot(om_test_var_stats_df)