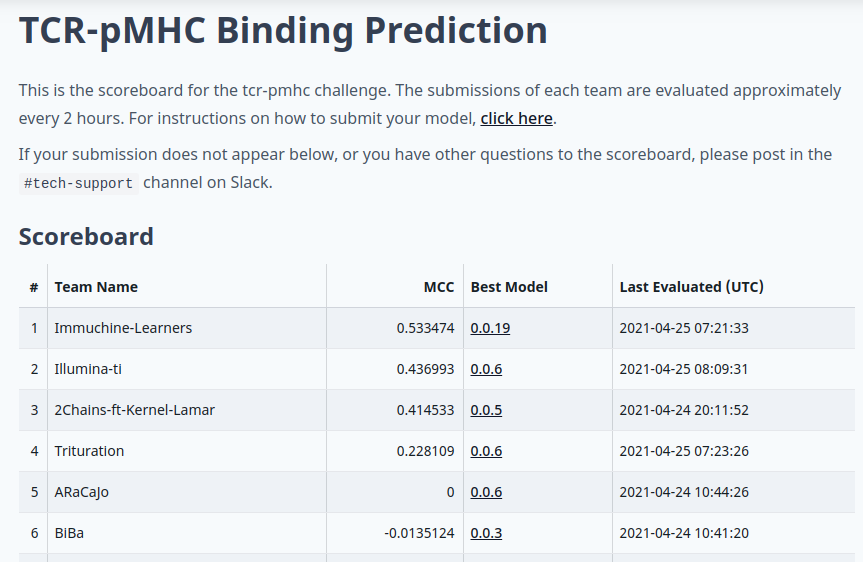
This challenge focuses on the binding between a T cell and its targets. The ability to predict this accurately has important applications in vaccine development and cancer immunotherapy. Using a dataset consisting of sequences of T-cell receptors and peptide-MHCs, together with the interaction energy predicted from structural models, participants will develop machine learning algorithms to predict the binding between a T cell and potential targets.
At prediction time, you are given a zip file using the --input-zip argument.
For each *.npz file, output your prediction.
The training dataset can be found in the data/training_set.zip file, and should be extracted to the data/train folder. An example CNN model is available as a notebook and in src directory.
You code should output a file called predictions.csv in the following format:
index,prediction
0,1
1,0
2,0
The continuous integration script in .github/workflows/ci.yml will automatically build the Dockerfile on every commit to the main branch. This docker image will be published as your hackathon submission to https://biolib.com/<YourTeam>/<TeamName>. For this to work, make sure you set the BIOLIB_TOKEN and BIOLIB_PROJECT_URI accordingly as repository secrets.
To read more about the benchmarking system click here.