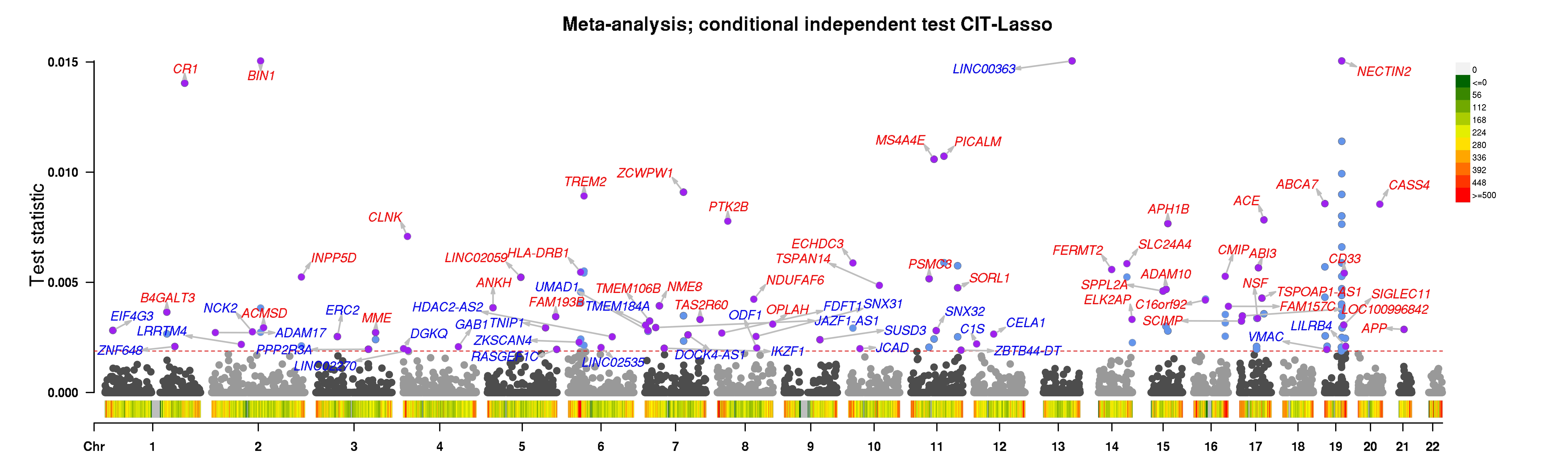
In a series of three papers, we show how the GhostKnockoff framework enables a knockoff-based analysis of GWAS summary statistics data with enhanced power compared to standard GWAS pipelines (e.g. LMM + SuSiE). The main methodology is described in the following papers
- Controlled Variable Selection from Summary Statistics Only? A Solution via GhostKnockoffs and Penalized Regression by Chen et al, with source code available under the chen_et_al folder.
- Second-order group knockoffs with applications to GWAS by Chu et al, with source code available in the chu_et_al folder.
- Beyond guilty by association at scale: searching for causal variants on the basis of genome-wide summary statistics by He et al, with source code available under the he_et_al folder.
For questions regarding this repo, please reach out to Zihuai He (zihuai@stanford.edu), Benjamin Chu (bbchu@stanford.edu), or Zhaomeng Chen (zc313@stanford.edu).