This repository contains the source code of KINference: Data-Driven Inference of Kinase Interaction Networks alongside two examplery runs. Refer to the GitHub Evaluation Repository for the evaluation code and the plotting results.
- Create a conda environment (mamba is recommended):
mamba create -n 'KINference' -c conda-forge r-base python
mamba activate KINference
- Install
pcst_fast
pip install pcst_fast
- Install required R packages (in an R terminal):
install.packages(c('tidyverse', 'data.table', 'BiocManager', 'devtools', 'argparse'))
devtools::install_github("evocellnet/funscoR")
BiocManager::install(c('OmnipathR', 'UniProt.ws'))
KINference has to be executed from the command line and within the git base directory via:
- Matrix based input:
Rscript run_KINference.R --x0.path='./data/example_data/example_intensities_x0.tsv' --x1.path='./data/example_data/example_intensities_x1.tsv' --output.path='example_run_results' --output.id='example_cond_run' --species='Human'
- Vector based input:
Rscript run_KINference.R --f.path='./data/example_data/example_intensities_f.tsv' --output.path='example_run_results' --output.id='example_cond_run' --species='Human'
--x1.path: Path to intensity data of condition 1 (default: NA).--x0.path: Path to intensity data of condition 0 (default: NA).--f.path: Path to log2-transformed intensities (default: NA).--output.path: Path to store the output (default: 'results').--output.id: Output identifier (default: 'key').--species: Species name (default: 'Human').--paired_samples: Bool variable that indicates if the samples are paired. Influences the computation of the log2-transformed intensities. See the paper for a more detailed explanation (default: FALSE).--alpha: Threshold for the baseline KIN percentile scoring (default: 0.9).--n: Threshold for the number of edges in the baseline KIN (default: 15).--beta: Threshold for the FS filter (default: 0.4).--gamma: Threshold for the DIFF filter (default: 1.0)--delta: Threshold for the CORR filter (default: 0.8)--epsilon: Threshold for the significance test of the CORR filter (default: 0.05)--m: Threshold for the number of samples needed for correlation calculation (default: 10).
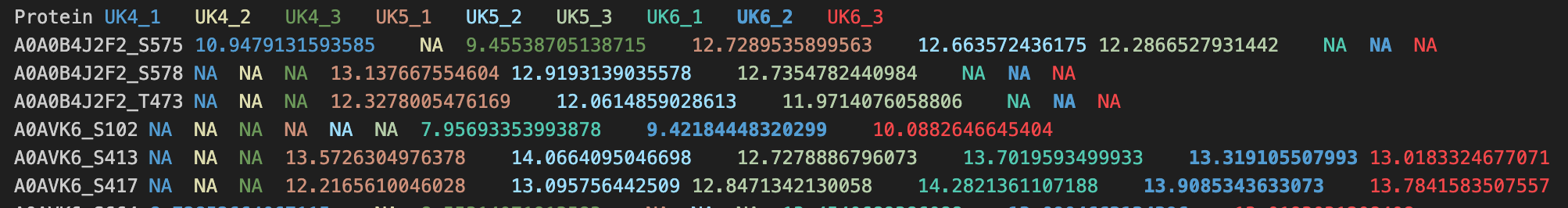
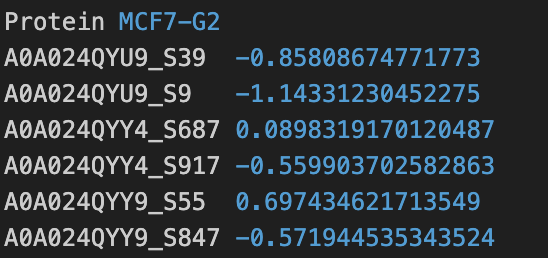
KINference can be run with either 2 input matrices of intensity measurements of phosphorylation sites or 1 input vector of log2FC-transformed intensities. The inputs have to be provided as tab-separated files. The matrices and the vector must contain one column named Protein. This column contains the annotations of the UniprotIDs and the phosphorylated amino acid (AA) and its sequence position (Pos) in UniprotID_AAPos format. See the screenshot for an example.
- Matrix input format: