| output |
|---|
github_document |
The goal of ggVolcano is to help users make a beautiful volcano map more easily, including general volcano plot(ggvolcano), gradient color volcano plot(gradual_volcano) and GO term volcano plot(term_volcano).
You can install the development version of ggVolcano like so:
# install.packages("devtools")
devtools::install_github("BioSenior/ggVolcano")- Make sure you have a DEG result data containing information on differentially expressed genes (including GeneName, Log2FC, pValue, FDR).
- If your data don't have a column named ‘regulate’, you can use the
add_regulatefunction to add. - Use function
ggvolcanoto make a general volcano plot. You can use?ggvolcanoto see the parameters of the function.
library(ggVolcano)
## basic example code
# load the data
data(deg_data)
# use the function -- add_regulate to add a regulate column
# to the DEG result data.
data <- add_regulate(deg_data, log2FC_name = "log2FoldChange",
fdr_name = "padj",log2FC = 1, fdr = 0.05)
# plot
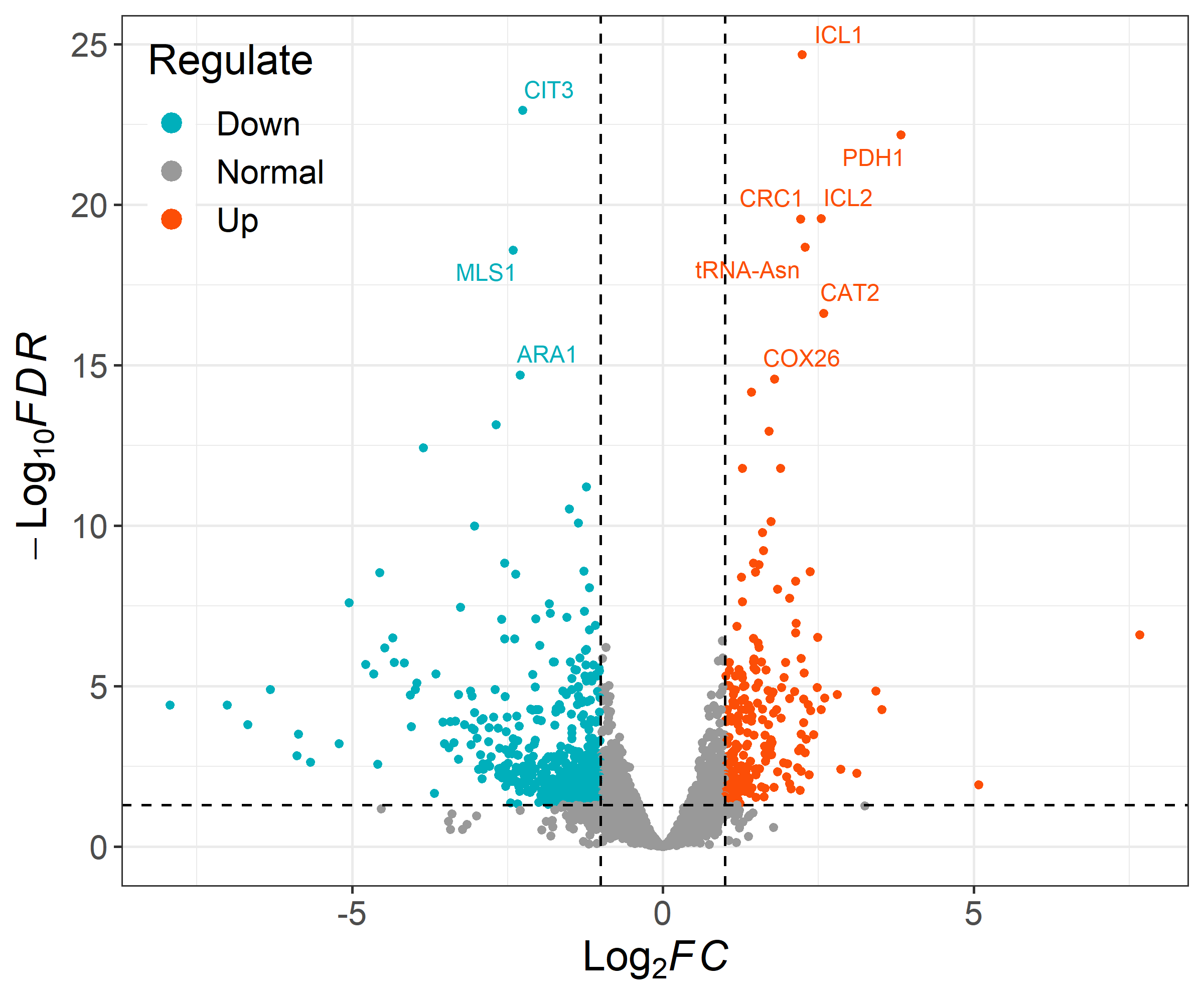
ggvolcano(data, x = "log2FoldChange", y = "padj",
label = "row", label_number = 10, output = FALSE)
- You must set x and y to the corresponding column names in your data.
- If you want to add label to the points, please set label to the corresponding column names in your data. And the label_number parameter can adjust the number of displayed labels.(Select the top label_number genes in terms of FDR value.)
# Change the fill and color manually:
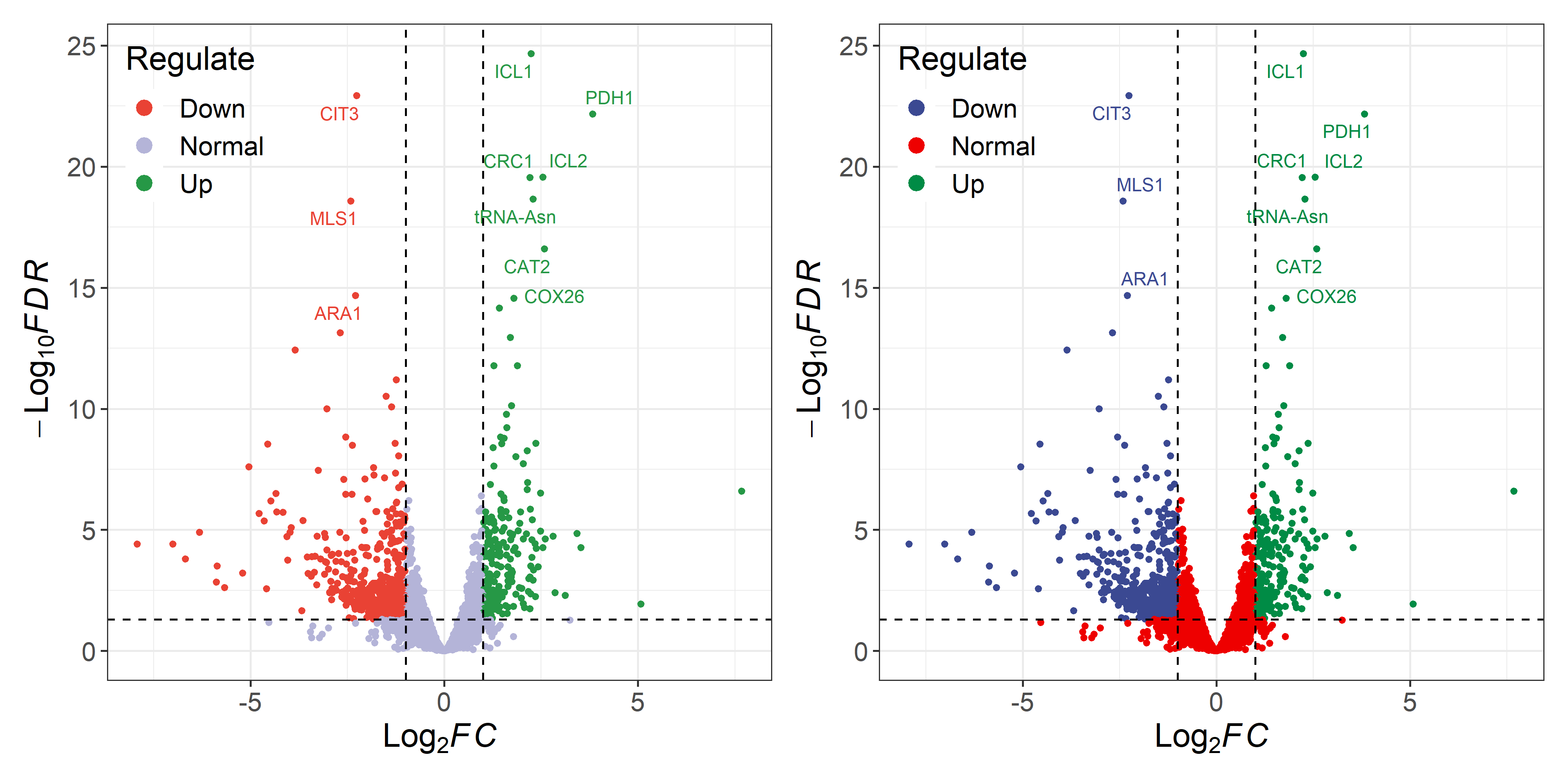
p1 <- ggvolcano(data, x = "log2FoldChange", y = "padj",
fills = c("#e94234","#b4b4d8","#269846"),
colors = c("#e94234","#b4b4d8","#269846"),
label = "row", label_number = 10, output = FALSE)
p2 <- ggvolcano(data, x = "log2FoldChange", y = "padj",
label = "row", label_number = 10, output = FALSE)+
ggsci::scale_color_aaas()+
ggsci::scale_fill_aaas()
#> Scale for 'colour' is already present. Adding another scale for 'colour', which
#> will replace the existing scale.
#> Scale for 'fill' is already present. Adding another scale for 'fill', which
#> will replace the existing scale.
library(patchwork)
p1|p2- Make sure you have a DEG result data containing information on differentially expressed genes (including GeneName, Log2FC, pValue, FDR).
- If your data don't have a column named ‘regulate’, you can use the
add_regulatefunction to add. - Use function
ggvolcanoto make a general volcano plot. You can use?gradual_Volcanoto see the parameters of the function.
# plot
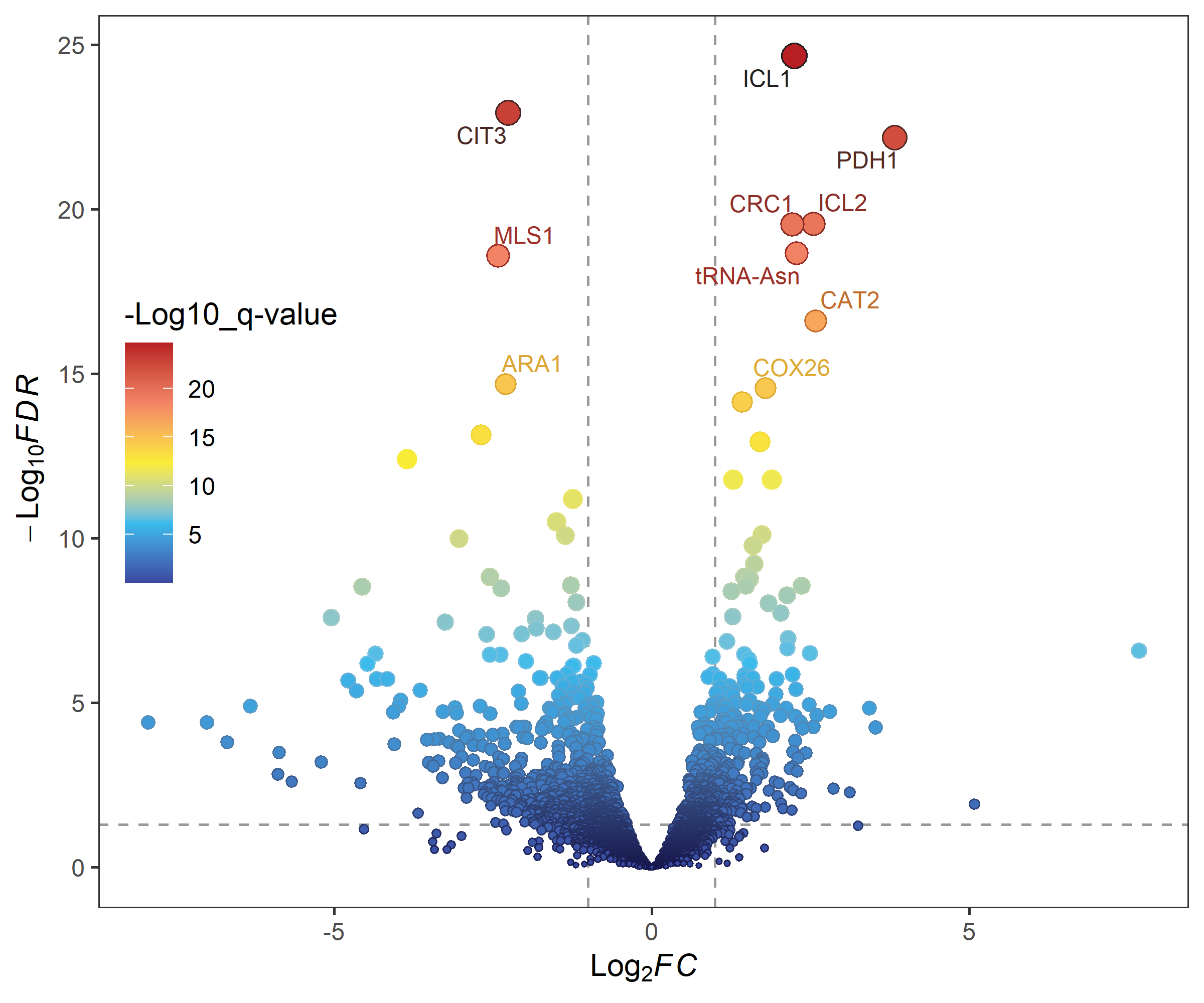
gradual_volcano(deg_data, x = "log2FoldChange", y = "padj",
label = "row", label_number = 10, output = FALSE)library(RColorBrewer)
# Change the fill and color manually:
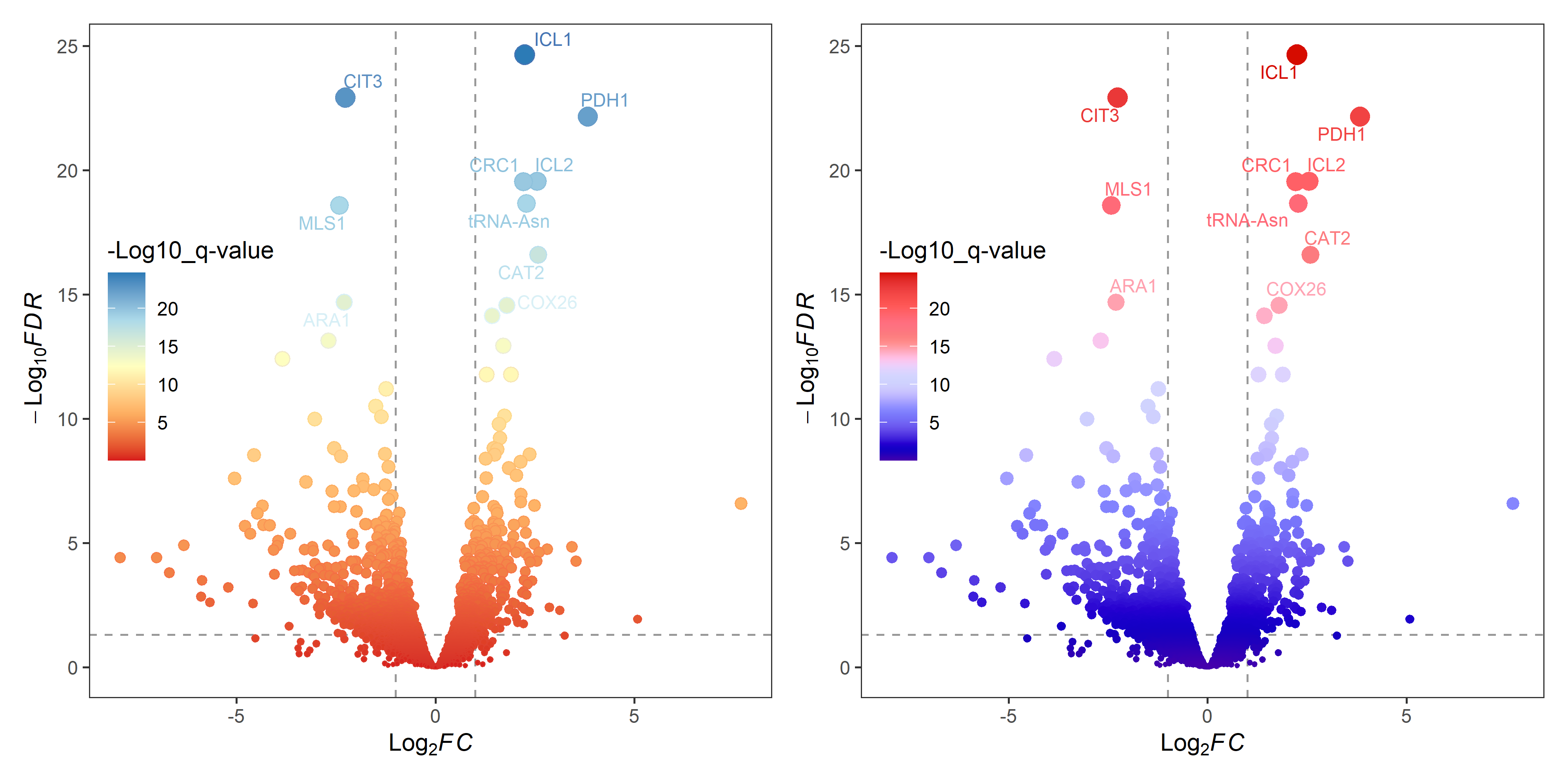
p1 <- gradual_volcano(data, x = "log2FoldChange", y = "padj",
fills = brewer.pal(5, "RdYlBu"),
colors = brewer.pal(8, "RdYlBu"),
label = "row", label_number = 10, output = FALSE)
p2 <- gradual_volcano(data, x = "log2FoldChange", y = "padj",
label = "row", label_number = 10, output = FALSE)+
ggsci::scale_color_gsea()+
ggsci::scale_fill_gsea()
#> Scale for 'colour' is already present. Adding another scale for 'colour', which
#> will replace the existing scale.
#> Scale for 'fill' is already present. Adding another scale for 'fill', which
#> will replace the existing scale.
library(patchwork)
p1|p2
- If you want to adjust the size range of the points, you can use
pointSizeRange = c(min_size,max_size).
- Make sure you have a DEG result data containing information on differentially expressed genes (including GeneName, Log2FC, pValue, FDR).
- Except a DEG result data, you also need a term data which is a two columns dataframe containing some genes' GO terms information.
- If your data don't have a column named ‘regulate’, you can use the
add_regulatefunction to add. - Use function
ggvolcanoto make a general volcano plot. You can use?term_Volcanoto see the parameters of the function.
data("term_data")
# plot
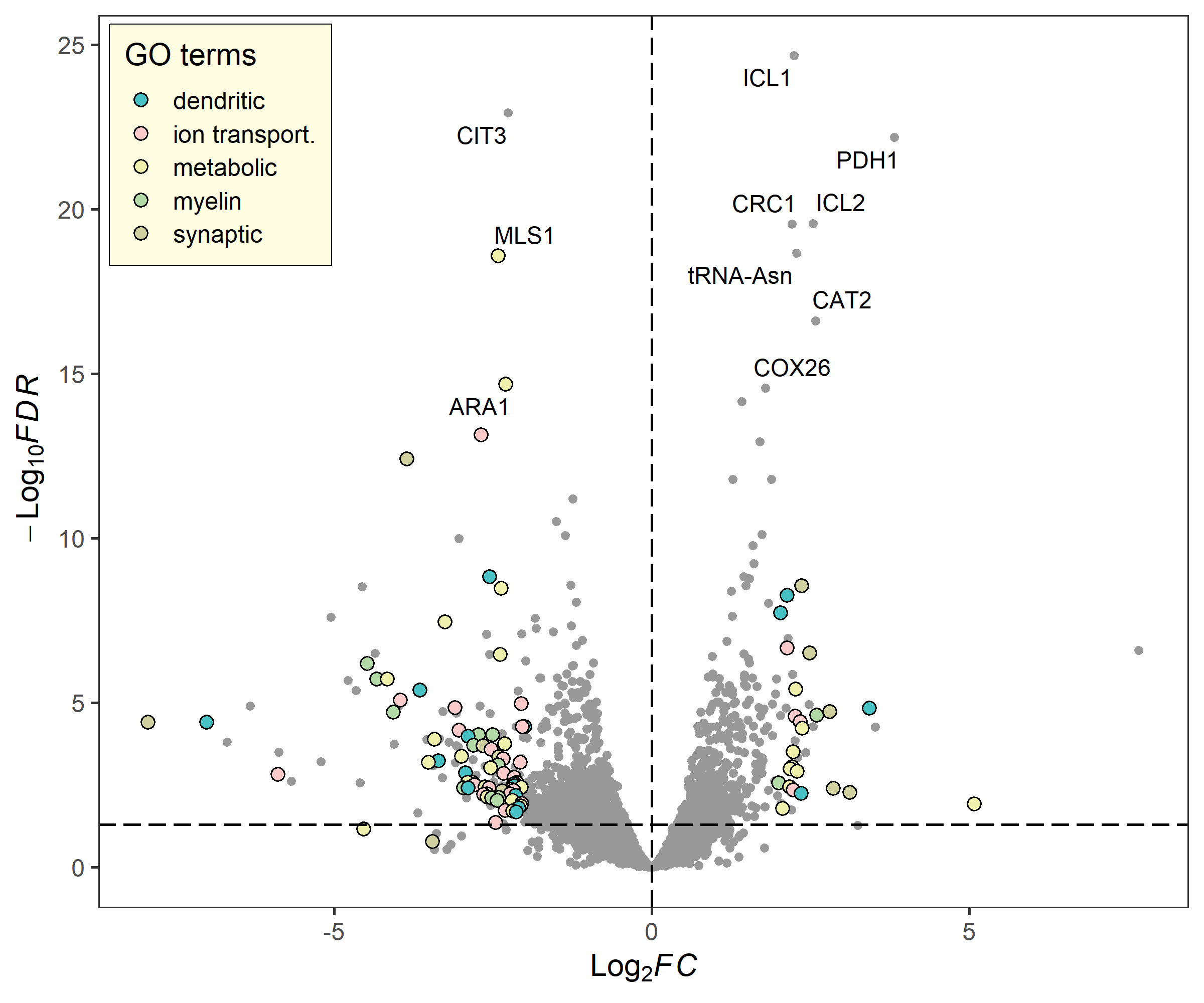
term_volcano(deg_data, term_data,
x = "log2FoldChange", y = "padj",
label = "row", label_number = 10, output = FALSE)library(RColorBrewer)
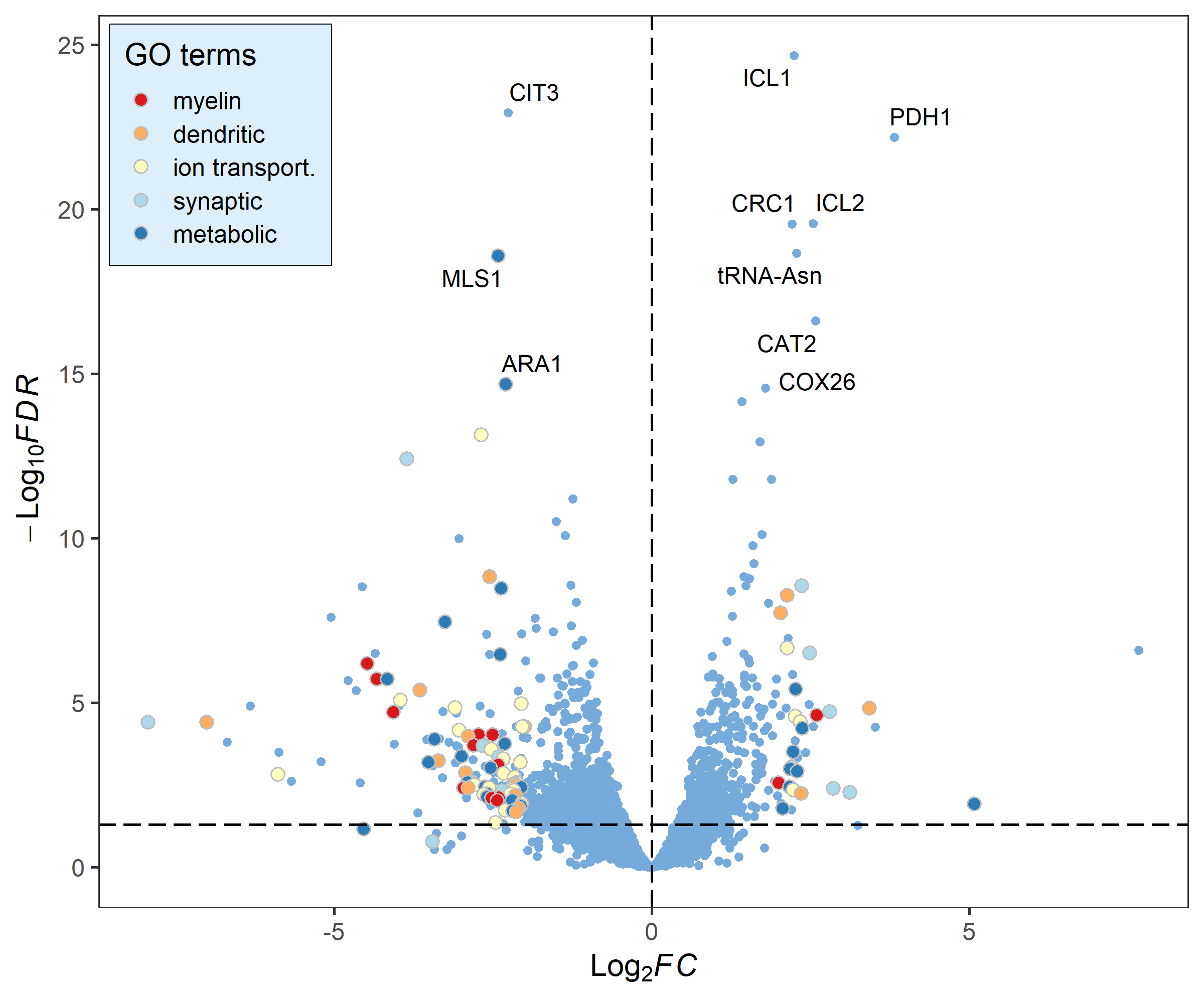
# Change the fill and color manually:
deg_point_fill <- brewer.pal(5, "RdYlBu")
names(deg_point_fill) <- unique(term_data$term)
term_volcano(data, term_data,
x = "log2FoldChange", y = "padj",
normal_point_color = "#75aadb",
deg_point_fill = deg_point_fill,
deg_point_color = "grey",
legend_background_fill = "#deeffc",
label = "row", label_number = 10, output = FALSE)Welcome to pay attention to the BioSenior to get more practical tutorials on scientific mapping!