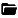An educational tool to get hands-on experience with the k-space and the effects of various modifications on the resulting image after an inverse Fourier transform.
K-space Explorer is written in Python 3 and uses open source libraries, so it can be used for free and the source code can be inspected to peek under the hood.
To try some features of the desktop app, you can now visit the K-Space Explorer Online
(this version is also on GitHub)
The desktop app has many useful features, such as:
- A modern responsive user interface using Qt
- Automatic Fourier transform to instantaneously visualise changes
- Load your own images and analyse artefacts originating from k-space
- Short explanation for various functions within the software
☕ This app was created in my spare time. If you find it useful please consider buying me a coffee with PayPal ☕
- Installation
- Starting the program
- Usage
- Troubleshooting
- Comparison to Other Similar Projects
- Latest changes
- Known bugs
- Planned features
- Disclaimer and limitations
- Author & Contributors
- References
-
You will need to have the following software and packages
- Python 3 (ideally the latest version). Download from the Python 3 homepage.
-
Required Packages for Python 3:
- PyQt5 - provides graphical user interface
- Pillow - opens regular images such as jpg or png
- NumPy - handles FFT transforms and array operations
- pydicom - DICOM format medical image reader
Install via pip by copying the command below to a command prompt (Windows:
Win+Rand typecmdand press Enter)pip3 install numpy pydicom Pillow PyQt5
-
Download the app and extract it
Navigate to the folder that contains the extracted software and run it by typing the command below
python kspace.pyTo save you navigating on Windows, just open the folder where the app was extracted and:
- Click the address bar (the line that looks something like this: "This PC > Downloads > kspace-explorer-master")
- Delete the address and type
cmdand press Enter - A window should appear, and you can now type
python kspace.py
KSE automatically loads a default image, but you can easily switch images by either:
You can also load a prepared raw data file to view a multichannel image acquisition. Please refer to the relevant section for instructions.
 |
|---|
| A real-life herringbone artefact and the corresponding k-space |
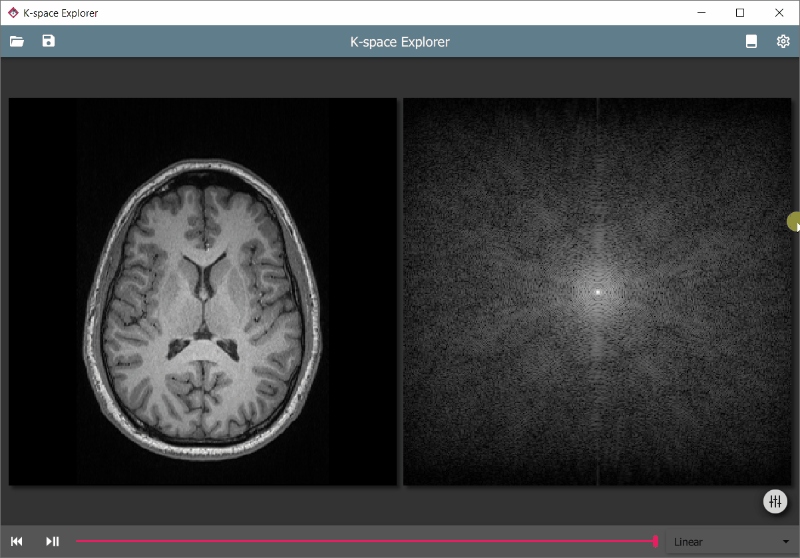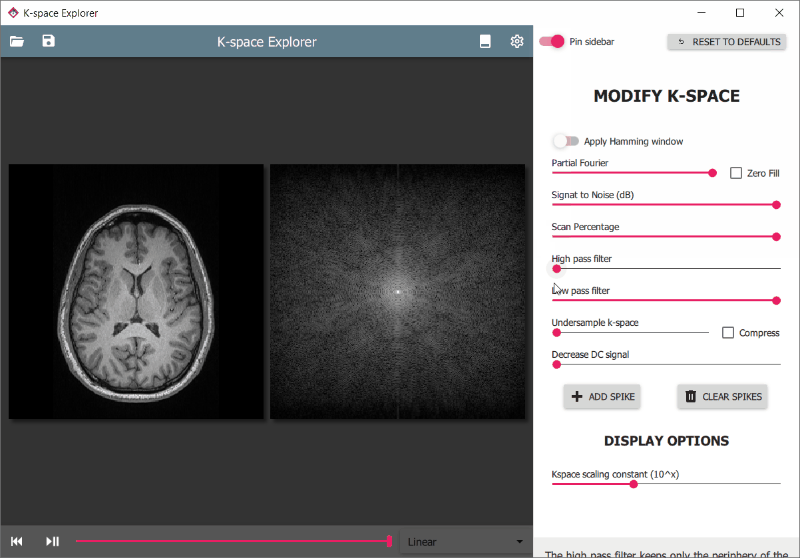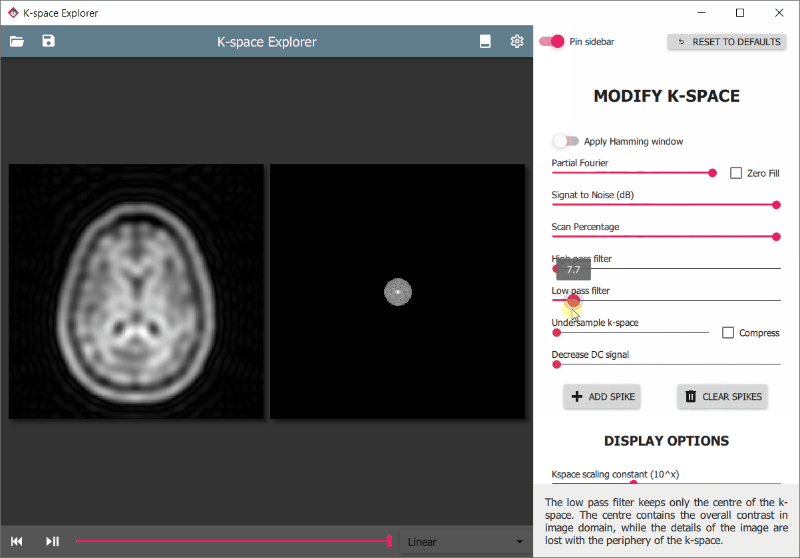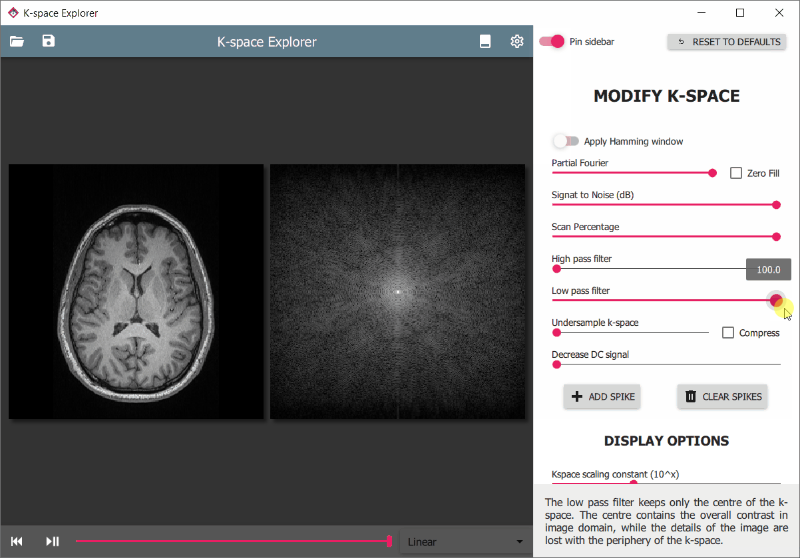
There are various modifiers available to edit the k-space and see the effects on the resulting image. These are accessible from the drawer panel on the right. To access it:
- Click and Drag inwards from the right side of the window
- Press the
Tabkey - Click the round button
 on the lower right side
on the lower right side
You can use the controls in the footer. The footer can be toggled by using the toolbar icon  or by hitting
or by hitting F7
- To start or continue the acquisition, press Play/Pause
 or press
or press F5 - To rewind press Rewind
 or press
or press F4 - You can change the simulation mode with the dropdown box on right-hand side
Your modified images can be saved to your computer by either
Then select the location and the filename. Visual representation of the k-space and the corresponding image will be saved with _k and _i suffixes, respectively.
Please note that if you select the tiff format, k-space image will be saved with 32-bit depth. This not handled well with many image viewers.
To enhance certain parts of the image for viewing it is often useful to change the brightness or contrast of the displays.
- Hold the right mouse button and move up/down to change image or k-space brightness
- Hold the right mouse button and move left/right to change image contrast
With windowing it is possible to limit the displayed image pixel intensity range.
- Drag mouse left/right with middle mouse button pressed to change window width
- Drag mouse up/down with middle mouse button pressed to change window centre
K-space Explorer can load a specially prepared (simplified) raw data file, which can contain data from multiple coil elements.
An example file can be found in the images folder (obtained from Harvard SMURF public domain repository [1]).
The raw data file can be loaded just like a normal image - drag & drop or using the Open New Image dialog.
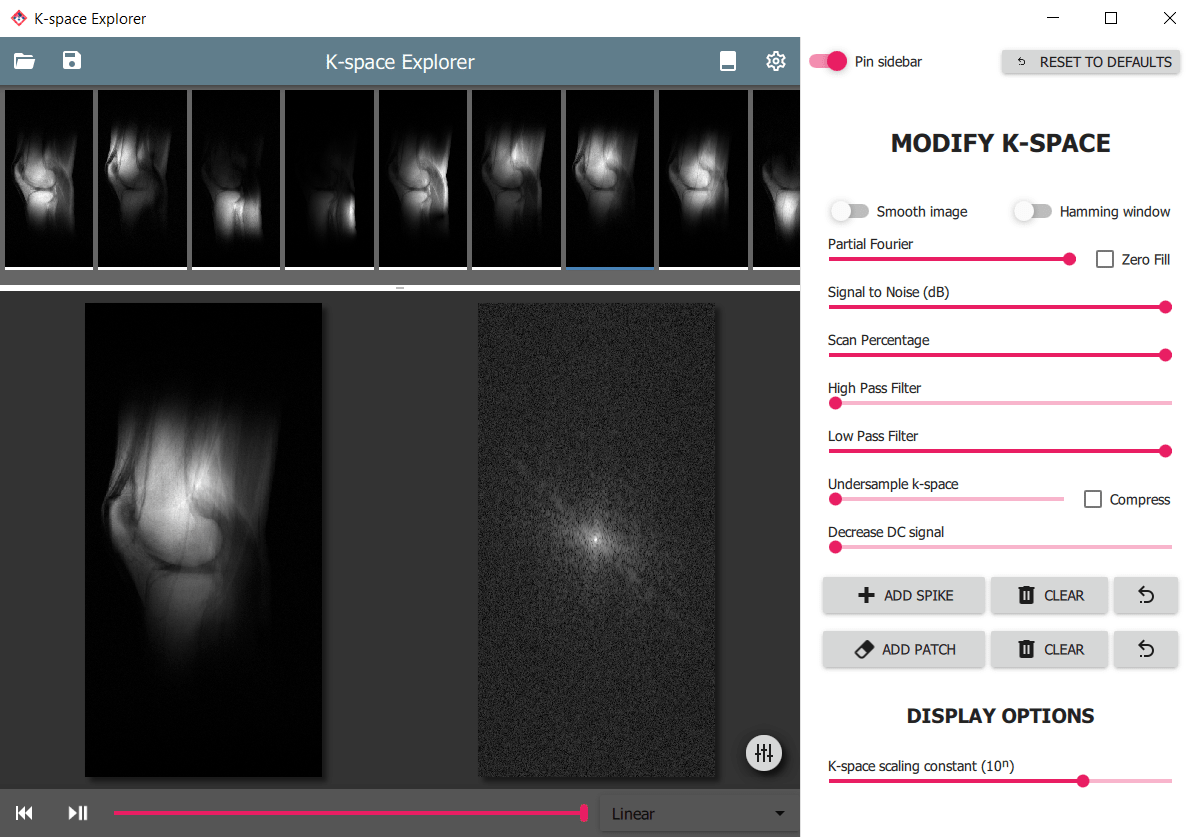 |
|---|
| Multi-channel raw data viewer interface of K-space Explorer |
The raw data file is a simple 3D numpy array with [channel, rows, columns] arrangement. If the array is prepared in Python it can be saved to .npy file with the numpy.save function.
Raw data can be obtained from the scanner with differing methods and proprietary formats determined by the scanner vendor. Siemens scanner raw data can be exported using this tutorial. The .dat file can be processed in Python using the twixtools package from the German Center for Neurodegenerative Diseases within the Helmholtz Association (DZNE).
See the example script below on how to read the image data and save it to the format K-space explorer can handle.
import numpy as np
import twixtools
filename = "knee_TSE_V2_waterSat.dat"
save_as = "knee_kspace_explorer"
def import_kspace(mdb_list):
# Function taken from twixtools documentation
image_mdbs = []
for mdb in mdb_list:
if mdb.is_image_scan():
image_mdbs.append(mdb)
n_line = 1 + max([mdb.cLin for mdb in image_mdbs])
n_part = 1 + max([mdb.cPar for mdb in image_mdbs])
n_channel, n_column = image_mdbs[0].data.shape
out = np.zeros([n_part, n_line, n_channel, n_column], dtype=np.complex64)
for mdb in image_mdbs:
out[mdb.cPar, mdb.cLin] += mdb.data
return out
twix_data = import_kspace(twixtools.read_twix(filename)[-1]['mdb'])
extract_part0 = np.zeros((twix_data.shape[1:4]), dtype=np.dtype(np.complex64))
extract_part0[:, :, :] = twix_data[0, :, :, :]
# Reorder axis to [channel, row, column]
array_to_save = np.transpose(extract_part0, (1, 0, 2))
# Optional - rotate all channels by 90 degress
# array_to_save = np.rot90(array_to_save, 3, (1, 2))
np.save(save_as, array_to_save, allow_pickle=False)If the app does not behave as expected, you can post issues here with your GitHub account.
To help diagnose the problem, please try and run K-space Explorer with logging enabled.
To enable logging mode, run the app with the --log command line argument.
python kspace.py --logThe file kspace.log will be populated with debugging information.
You can then upload the contents of the file here
and include a link to the published log in your issue.
This app was directly influenced by the article of D. Moratal et al. [2], however my aim was to go beyond the functionality that it offers. Several similar software is available for different computing environments. Here is a non-exhaustive list of them:
- k-Space Tutorial (PC, Matlab) [2]
- Journey through k-space (PC, Matlab) [3]
- A k-Space Odyssey (iOS) [4]
- K-Spapp (Android, free) [5]
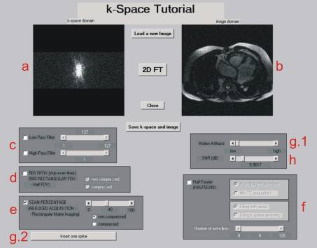 |
|---|
| A screenshot from k-Space Tutorial by D. Moratal et al. |
Matlab is a requirement for many similar apps. Matlab is proprietary software and can be costly to purchase a license. K-space Explorer only uses free software to make it more accessible.
The aim of K-space Explorer is to provide a smooth, modern UI with familiar tools and instant response whenever possible. Updates happen real time so immediate feedback is given for the effect of the changes.
To get a deeper understanding of the inner workings the code can be inspected. In-line documentation can help in the understanding of the mathematical principles behind various interactions.
- Basic logging
- View raw data with images from a multichannel acquisition
- RGB DICOMs are not supported
- Accelerated scanning method simulation (SENSE, GRAPPA, POCS)
- Multiple languages
- CLAHE enhancement
This software is not intended for medical use. Even if a scanner original DICOM file is used, the resulting k-space is not equivalent to the scanner raw data as it contains all post-acquisition modifications applied by the scanner software.
 |
|---|
| Gergely Biró |
[1] Bachrata, Beata, 2020, "SMURF (raw MRI data)", https://doi.org/10.7910/DVN/XNMCYI, Harvard Dataverse, V3
[2] Moratal, D., Vallés-Luch, A., Martí-Bonmati, L., & Brummers, M. E. (2008). k-Space tutorial: An MRI educational tool for a better understanding of k-space. Biomedical Imaging and Intervention Journal, 4(1). https://doi.org/10.2349/biij.4.1.e15
[3] Qureshi, M., Kaleem, M., & Omer, H. (2017). Journey through k-space: An interactive educational tool. Biomedical Research (India), 28(4), 1618–1623.
[4] Ridley, E. L. (21/03/2017). Mobile App Spotlight: A k-Space Odyssey. Source: AuntMinnie.com: https://www.auntminnie.com/index.aspx?sec=sup&sub=mri&pag=dis&ItemID=116900&wf=7612
[5] K-Spapp - https://mrapps.jouwweb.nl/