Learning protein structural similarity from sequence alone.
DeepBLAST can be installed from pip via
pip install deepblast
To install from the development branch run
pip install git+https://github.com/flatironinstitute/deepblast.git
The pretrained DeepBLAST model can be downloaded here.
The TM-align structural alignments used to pretrain DeepBLAST can be found here
See the Malisam and Malidup websites to download their datasets.
We have 2 command line scripts available, namely deepblast-train and deepblast-eval.
deepblast-train takes in as input a tab-delimited format of with columns
query_seq_id | key_seq_id | tm_score1 | tm_score2 | rmsd | sequence1 | sequence2 | alignment_string
See an example here of what this looks like. At this moment, we only support parsing the output of TM-align. The parsing script can be found under
deepblast/dataset/parse_tm_align.py [fname] [output_table]
Once the data is configured and split appropriately, deepblast-train can be run.
The command-line options are given below (see deepblast-train --help for more details).
usage: deepblast-train [-h] [--gpus GPUS] [--grad-accum GRAD_ACCUM] [--grad-clip GRAD_CLIP] [--nodes NODES] [--num-workers NUM_WORKERS] [--precision PRECISION] [--backend BACKEND]
[--load-from-checkpoint LOAD_FROM_CHECKPOINT] --train-pairs TRAIN_PAIRS --test-pairs TEST_PAIRS --valid-pairs VALID_PAIRS [--embedding-dim EMBEDDING_DIM]
[--rnn-input-dim RNN_INPUT_DIM] [--rnn-dim RNN_DIM] [--layers LAYERS] [--loss LOSS] [--learning-rate LEARNING_RATE] [--batch-size BATCH_SIZE]
[--multitask MULTITASK] [--finetune FINETUNE] [--mask-gaps MASK_GAPS] [--scheduler SCHEDULER] [--epochs EPOCHS]
[--visualization-fraction VISUALIZATION_FRACTION] -o OUTPUT_DIRECTORY
This will evaluate how much the deepblast predictions agree with the structural alignments.
The deepblast-train command will automatically evaluate the heldout test set if it completes.
However, a separate deepblast-evaluate command is available in case the pretraining was interrupted. The commandline options are given below (see deepblast-evaluate --help for more details)
usage: deepblast-evaluate [-h] [--gpus GPUS] [--num-workers NUM_WORKERS] [--nodes NODES] [--load-from-checkpoint LOAD_FROM_CHECKPOINT] [--precision PRECISION] [--backend BACKEND]
--train-pairs TRAIN_PAIRS --test-pairs TEST_PAIRS --valid-pairs VALID_PAIRS [--embedding-dim EMBEDDING_DIM] [--rnn-input-dim RNN_INPUT_DIM]
[--rnn-dim RNN_DIM] [--layers LAYERS] [--loss LOSS] [--learning-rate LEARNING_RATE] [--batch-size BATCH_SIZE] [--multitask MULTITASK]
[--finetune FINETUNE] [--mask-gaps MASK_GAPS] [--scheduler SCHEDULER] [--epochs EPOCHS] [--visualization-fraction VISUALIZATION_FRACTION] -o
OUTPUT_DIRECTORY
import torch
from deepblast.trainer import LightningAligner
from deepblast.dataset.utils import pack_sequences
from deepblast.dataset.utils import states2alignment
import matplotlib.pyplot as plt
import seaborn as sns
# Load the pretrained model
model = LightningAligner.load_from_checkpoint(your_model_path)
# Load on GPU (if you want)
model = model.cuda()
# Obtain hard alignment from the raw sequences
x = 'IGKEEIQQRLAQFVDHWKELKQLAAARGQRLEESLEYQQFVANVEEEEAWINEKMTLVASED'
y = 'QQNKELNFKLREKQNEIFELKKIAETLRSKLEKYVDITKKLEDQNLNLQIKISDLEKKLSDA'
pred_alignment = model.align(x, y)
x_aligned, y_aligned = states2alignment(pred_alignment, x, y)
print(x_aligned)
print(pred_alignment)
print(y_aligned)
x_ = torch.Tensor(model.tokenizer(str.encode(x))).long()
y_ = torch.Tensor(model.tokenizer(str.encode(y))).long()
# Pack sequences for easier parallelization
seq, order = pack_sequences([x_], [y_])
seq = seq.cuda()
# Predict expected alignment
A, match_scores, gap_scores = model.forward(seq, order)
# Display the expected alignment
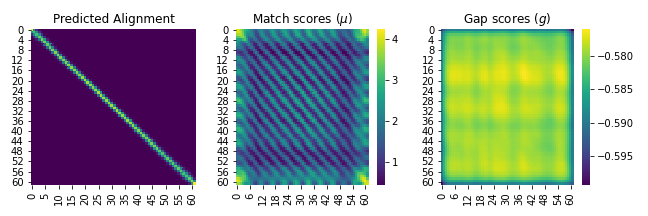
fig, ax = plt.subplots(1, 3, figsize=(9, 3))
sns.heatmap(A.cpu().detach().numpy().squeeze(), ax=ax[0], cbar=False, cmap='viridis')
sns.heatmap(match_scores.cpu().detach().numpy().squeeze(), ax=ax[1], cmap='viridis')
sns.heatmap(gap_scores.cpu().detach().numpy().squeeze(), ax=ax[2], cmap='viridis')
ax[0].set_title('Predicted Alignment')
ax[1].set_title('Match scores ($\mu$)')
ax[2].set_title('Gap scores ($g$)')
plt.tight_layout()
plt.show()The output will look like
IGKEEIQQRLAQFVDHWKELKQLAAARGQRLEESLEYQQFVANVEEEEAWINEKMTLVASED
::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
QQNKELNFKLREKQNEIFELKKIAETLRSKLEKYVDITKKLEDQNLNLQIKISDLEKKLSDA
Q : How do I interpret the alignment string?
A : The alignment string is used to indicate matches and mismatches between sequences. For example consider the following alignment
ADQSFLWASGVI-S------D-EM--
::::::::::::2:222222:2:122
MHHHHHHSSGVDLWSHPQFEKGT-EN
The first 12 residues in the alignment are matches. The last 2 characters indicate insertions in the second sequence (hence the 2 in the alignment string), and the 3rd to last character indciates an insertion in the first sequence (hence the 1 in the aligment string).