Runs an sRNA-seq data analysis pipeline on a collection of fastq.gz, fastq, fq, or fq.gz files.
- sRNA Snakemake Workflow
- Pipeline Ins and Outs
- Run Within A Container
- About the Output Files
- Flowchart of Pipeline Functions
- References
- Snakemake (tested with version 5.4.5)
- Trimgalore (tested with version 0.6.2)
- cutadapt (tested with version 2.3)
- fastqc (tested with version 0.11.7)
- samtools (tested with version 1.9)
- bowtie (tested with version 1.2.2)
- ShortStack (tested with version 3.8.5)
- RNAfold (tested with version 2.3.2)
- XZ Utils] (tested with version 5.2.2)
- liblzma (tested with version 5.2.2)
-
Ensure that the Dependencies for the pipeline have been installed.
-
Clone this repository into your current directory from the command line by running:
$ git clone https://github.com/boseHere/sRNA_snakemake_workflow $ cd sRNA_snakemake_workflow -
Add your sample files to the
data/1_raw/directory. These can be fastq, fq, fastq.gz, or fq.gz files. See Samples for more info about this. -
Add your genomes files (chloroplast and mitochondria genome, reference genome, and optional non-coding RNA genome) to the corresponding subdirectories in the
genomesdirectory. See Genomes for more info about this. -
See Directory Structure for a concise glance at what your directory structure should look like from the top level before running the pipeline. Check that your directory structure within the
sRNA_snakemake_workflowfolder matches this. -
Fill out the config.yaml file in the top level directory. See Editing Config for more information on the different sections of the file and how to fill them out. The config.yaml file that is downloaded with this repository also contains comments to assist with filling it in.
-
Execute the following on the command line from the top level of the directory structure:
$ snakemake --cores # INSERT MAX NUMBER OF CORES HEREIf a previous snakemake process was interrupted, you may need to run the following command to unlock the directory before running the snakemake pipeline again:
$ snakemake --unlockEnsure you have the following directory structure in place before running snakemake from the top level directory. This structure should be already in place if you download this repository with git clone, and should only require that you fill in your sample and genome files.
./sRNA_snakemake_workflow
├── _data
│ └── 1_raw
│ └── # YOUR FASTQ.GZ, FASTQ, FQ.GZ, AND FQ FILES CONTAINING SRNA-SEQ DATA HERE
├── _genomes
│ └── _chloro_mitochondria
│ └── # MULTI-FASTA FILE CONTAINING CHLOROPLAST AND/OR MITOCHODRIAL GENOME(S) FOR YOUR ORGANISM HERE
│ └── _filter_rna
│ └── # MULTI-FASTA FILE CONTAINING NON-CODING RNA SEQUENCES TO BE FILTERED OUT OF INPUT READS HERE (OPTIONAL)
│ └── _reference_genome
│ └── # MULTI-FASTA FILE CONTAINING THE GENOME FOR YOUR ORGANISM HERE
├── _output_logs
├── _scripts
│ └── build_config.sh
│ └── fastq_readlength_profile.py
│ └── index_genomes.sh
│ └── match_qual_v2.py
│ └── rfam_sql.py
├── config.yaml
└── Snakefile
As snakemake runs, the data folder will become populated with folders numbered by the order they are created.
Editing the config.yaml file for this pipeline allows you to specify your files, reference genomes, software pathways, and trimming parameters. To edit the config file, run
$ nano config.yamlthen fill out the sections as described below. Save by pressing Ctrl-o + enter, then exit the config file via Ctrl-x.
You have the option to either fill in all sections of the config.yaml file manually, or to allow our custom script build_config.sh to fill in the names of your samples and genomes according to what you have in your /data/1_raw/ and /genomes/ directories.
This option was created because filling in the config.yaml with the names of your samples and genomes can be tedious, especially if you have a lot of samples or don't already have a list of all your sample filenames without their extensions. Further specifications on filling out the config file are noted in the config.yaml template that comes with the download of this pipeline, so feel free to fill it out manually if you prefer.
Otherwise, to have the config.yaml file automatically filled in with the names of your samples/genomes, simply run the following from the top level of the directory structure:
$ chmod +x ./scripts/build_config.sh
$ ./scripts/build_config.shAfter this, run:
$ nano config.yamlto proceed with filling in the Trimming, Aligning, Threads, and Paths sections of the config file (or use your text editor of choice). These sections are not filled in manually by the build_config.sh program, and must be specified by the user. The following sections will explain more about these specifications.
These config.yaml options are used by the Trim Galore program in the pipeline to size and quality select reads from the raw data files.
min_length : <int>- Set the minimum read length you are interested in. The advised default for sRNA analysis is 19.
max_length : <int>- Set the maxmimum read length you are interested in. The advised default for sRNA analysis is 26.
adapter_seq : <str>- Set the adaptor sequence used when creating the libraries. Some commonly used adapters:
- Illumina Adapter: AGATCGGAAGAGC
- Illumina sRNA Adapter: TGGAATTCTCGG
- Nextera Adapter: CTGTCTCTTATA The default is the Illumina sRNA adapter
- Set the adaptor sequence used when creating the libraries. Some commonly used adapters:
quality : <int>- Set the minumum Phred score for reads. Reads with quality lower than this will be removed. The default is 30.
This pipeline (currently) will not work on sequences that have already been adapter-trimmed.
These options are used by the ShortStack aligner to align reads to a reference genome.
multi_map_handler : <str>- Fill in the desired protocol to handle multi-mapping reads during the alignment process. The options for this, as described by the ShortStack documentation include n (none), r (random), u (unique- seeded guide), or f (fractional-seeded guide). The suggested default is u, which assigns multi-mapping reads to regions with higher levels of uniquely mapping reads.
sort_memory : <int>G- Fill in the desired amount of memory to be allocated for sorting bam files. The default for this is 20G, though you may want to increase this if you find the pipeline crashing during the clustering step, or if you have many large sample files.
no_mirna : <str>- Enter Y if you do NOT want to perform miRNA annotation during the alignment. Enter N if you DO want to include the (optional) miRNAs annotation stage in the alignment.
filter_rna_bowtie : <int>filter_c_m_bowtie : <int>shortstack_cluster : <int>mapped_reads_samtools : <int>fastqc_report : <int>
Set the number of threads for each program to run with. The advised defaults are 5 for bowtie, shortstack, and samtools, and 1 for fastqc, but these numbers can be scaled up or down given server limitations. It is advised not to go above 10 threads for each program, as this decreases the number of processes snakemake can run in parallel.
Note: You do not need to fill in this section if you are running this pipeline through the Singularity container.
trim_galore : <str>bowtie : <str>ShortStack : <str>samtools : <str>
Give absolute paths to the trim_galore, bowtie, ShortStack, and samtools software if they are not already sym-linked to a location in /usr/local/bin/. To test if these software are sym-linked, you can run the following on the command line.
$ which trim_galore
$ which bowtie
$ which ShortStack
$ which SamtoolsIf these lines return a path, leave this section as is upon downloading.
Give names of the sample files located in your /data/1_raw directory without file extensions.
Example
samples:
- sample_name1
- sample_name2
- sample_name3Sample names should be indented using 4 spaces (not the indent key), and be preceded by a dash character "-" and another space.
Fill in the three absolute paths with the names of your genome files.
Note: The noncoding RNA filtering step of this pipeline is recommended for sRNA alignment, but not required for the pipeline to run. This step removes contaminating commonly highly expressed RNAs from your sequences. The .fasta file, containing these contaminating sequences, used to perform this filtering step should be placed in /genomes/filter_rna/. However, if you choose to not use this step, then no file is required in this directory.
MAKE SURE that the file you use for this filtering step is stripped of microRNAs and preRNAs before running the pipeline; these should be included in the alignment process.
The genome file for this step can be obtained by running the following from the top level of the directory structure:
$ chmod +x ./scripts/rfam_sql.py
$ ./scripts/rfam_sql.py --output_dir ../genomes/filter_rna/ <ncbi taxonomy id of your organism>The NCBI taxonomy ID of your organism can be obtained from by searching your organism name here.
The three file paths require the BASE NAME of the genome file. This is simply the name of the genome files without the .fasta extensions. If you choose not to use the additional filtering step to remove additional contaminating RNAs, simply leave the file path as is.
Example (using the additional filtering step)
genomes:
filter_rna : ./genomes/filter_rna/my_rna_genome
chloro_mitochondria : ./genomes/chloro_mitochondria/my_cm_genome
reference_genome : ./genomes/reference_genome/my_ref_genomeExample (NOT using the additional filtering step)
genomes:
filter_rna : ./genomes/filter_rna/
chloro_mitochondria : ./genomes/chloroplast_mitocondrion/my_cm_genome
reference_genome : ./genomes/reference_genome/my_ref_genomeContainers are discrete, executable units of software that can be run as an isolated system, regardless of computing environment.
In the context of this pipeline, a container addresses potential issues with reproducibility and HPC compatability, in addition to allowing users to bypass the tedious process of installing all of the software dependencies.
With regards to reproducibility, as the software used in this pipeline continues to be updated by developers, it is possible that processes run by this pipeline may no longer be supported by one of these updates. This may prevent analysis from being reproducible in the future. The container solves this problem by preserving the necessary software versions that are known to support this pipeline.
Some HPC systems do not allow users to freely install software on the server, or may only host an out of date version of the software. However, most HPC systems support the running of containers through either Docker or Singularity.
We have put together a container as an alternative to installing all of the software dependencies independently. Running the pipeline using this container requires having either Docker or Singularity installed.
To pull the Docker image containing all the above software pre-installed into your current directory, run:
$ docker image pull bose1/mosher_lab_srna:ubuntu_18
If you are running the pipeline on a system with an older Linux kernel (which can often be the case on HPC systems), you may need to pull the Ubuntu 16.04 version of the container. This container will provide all the same software dependencies as the ubuntu 18.04 version of the container. To obtain this version of the container, run:
$ docker image pull bose1/mosher_lab_srna:ubuntu_16If you are running the pipeline using the Docker container, configure your sRNA Snakemake file directory and config.yaml file as directed in Pipeline Ins and Outs. Then run the following from the top level of the directory structure:
$ docker run mosher_lab_srna:ubuntu_18.sif snakemake --cores # INSERT MAX NUMBER OF CORES HERETo pull the singularity image containing all the above software pre-installed into your current directory, run:
$ singularity pull docker://bose1/mosher_lab_srna:ubuntu_18
If you are running the pipeline on a system with an older Linux kernel (which can often be the case on HPC systems), you may need to pull the Ubuntu 16.04 version of the container. This container will provide all the same software dependencies as the ubuntu 18.04 version of the container. To obtain this version of the container, run:
$ singularity pull docker://bose1/mosher_lab_srna:ubuntu_16Running either of these commands will download a .sif file. You will need to move this .sif file to the same location as the Snakefile.
If you are running the pipeline using the Singularity container, configure your sRNA Snakemake file directory and config.yaml file as directed in Pipeline Ins and Outs. Then run the following from the top level of the directory structure:
$ singularity exec mosher_lab_srna_ubuntu_18.sif snakemake --cores # INSERT MAX NUMBER OF CORES HEREOnce the pipeline has completed running, you will see 7 additional sub-directories appear in the /data/ directory alongside the 1_raw directory. These should include:
-
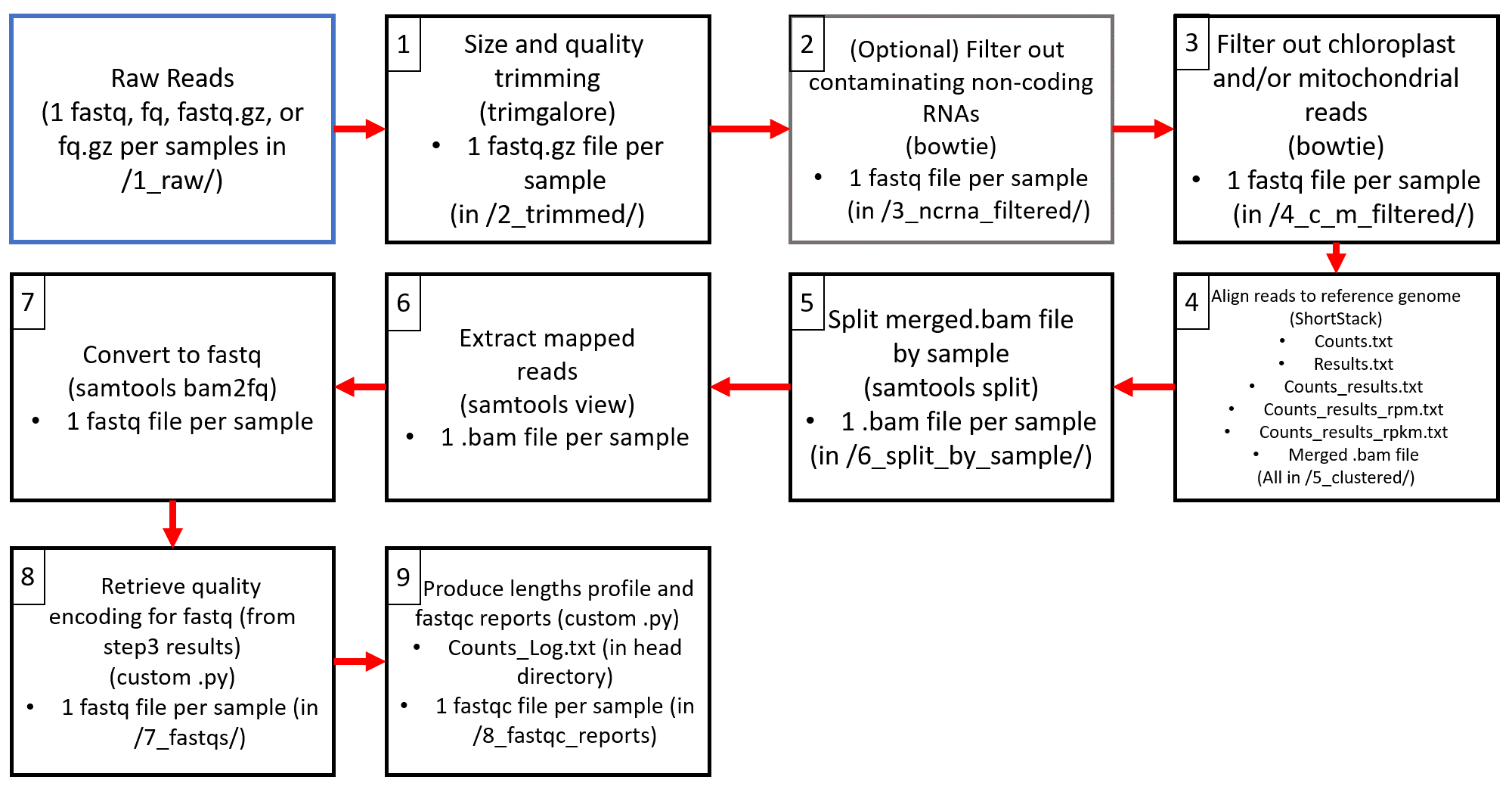
/2_trimmed/: This folder contains 1 fastq.gz file for each sample provided as input to the pipeline. The files in this folder have been selected for size and quality according to the specification given in the Trimming section of the config file.
-
/3_ncrna_filtered/: This folder will only appear if the additional filtering step, as described in Genomes was used. This folder will contain 1 fq.gz file for each sample provided as input to the pipeline. These files have had all contaminating non-coding RNAs filtered out.
-
/4_c_m_filtered/: This folder contains 1 fq.gz file for each sample provided as input to the pipeline. These files have had all chloroplast and/or mitochondrial reads filtered out.
-
/5_clustered/: This folder contains the output of aligning all sample files to the given reference genome using ShortStack. This includes:
- Counts.txt : Part of standard ShortStack output. See ShortStack documentation for further info about the contents of this file.
- counts_results.txt: Combines the alignment information from Counts.txt and Results.txt. Read counts are in the original raw count numbers. Excludes unplaced reads.
- counts_results_rpm.txt: Combines the alignment information from Counts.txt and Results.txt. Read counts are in reads per million mapped reads (RPM). Excludes unplaced reads.
- counts_results_rpkm.txt: Combines the alignment information from Counts.txt and Results.txt. Read counts are in reads per kilo base per million mapped reads (RPKM). Excludes unplaced reads.
- merged.bam: Alignment file. Part of standard ShortStack output. See ShortStack documentation for further info about the contents of this file and the ShortStack-specific tags used in these files.
- Results.txt: Part of standard ShortStack output. See ShortStack documentation for further info about the contents of this file.
- Unplaced.txt: Part of standard ShortStack output. See ShortStack documentation for further info about the contents of this file.
-
/6_split_by_sample/: This folder contains 1 bam file for each sample provided as input to the pipeline. This file contains the alignment information for each sample.
-
/7_fastqs/: This folder contains 1 fastq file for each sample provided as input to the pipeline. These files contain the reads that aligned to the reference genome.
This flowchart demonstrates the steps of the pipeline, including what tools are used, what files are created, and where they are stored.
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Cutadapt. DOI:10.14806/ej.17.1.200.
Grover JW, Kendall T, Baten A, Burgess D, Freeling M, King GJ, and Mosher RA. Maternal components of RNA ‐ directed DNA methylation are required for seed development in Brassica rapa. The Plant Journal. 2018. doi:10.1111/tpj.13910
Johnson NR, Yeoh JM, Coruh C, Axtell MJ. (2016). G3 6:2103-2111. doi:10.1534/g3.116.030452
Krueger, F. (2015). Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25.
R. Lorenz, S.H. Bernhart, C. Hoener zu Siederdissen, H. Tafer, C. Flamm, P.F. Stadler and I.L. Hofacker (2011), "ViennaRNA Package 2.0", Algorithms for Molecular Biology: 6:26