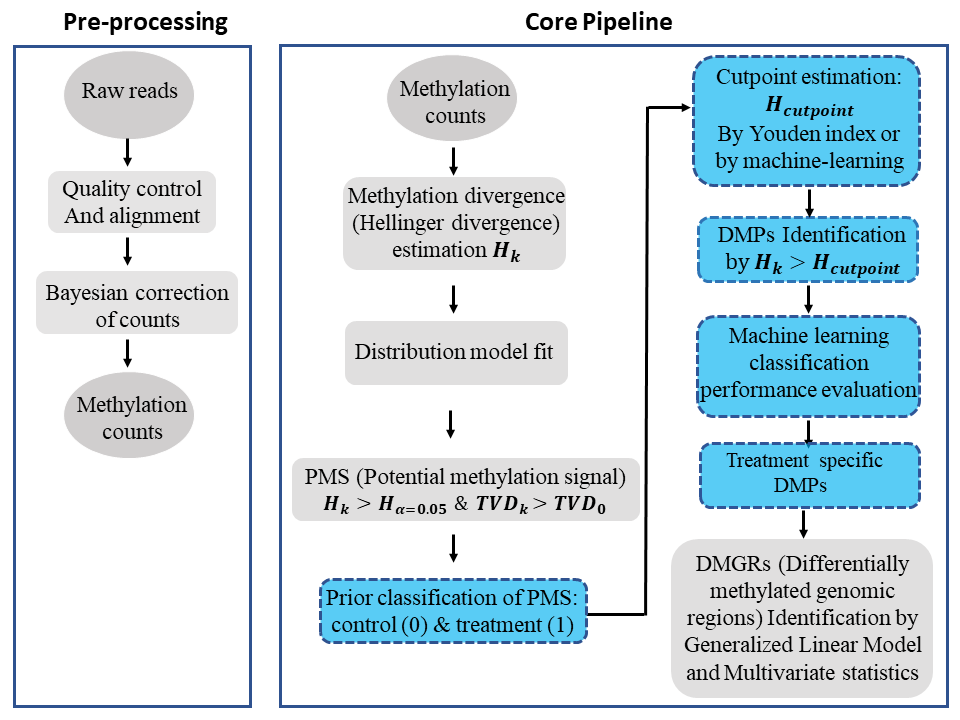
We have developed a novel methylome analysis procedure, Methyl-IT, based on signal detection and machine learning. Methylation analysis is confronted in Methyl-IT as a signal detection problem, and the method was designed to discriminate methylation regulatory signal from background noise induced by Brownian motion and thermal fluctuations. Our group has proposed an information thermodynamics approach to investigate genome-wide methylation patterning based on the statistical mechanical effect of methylation on DNA molecules (1-3). The information thermodynamics-based approach is postulated to provide greater sensitivity for resolving true signal from the thermodynamic background within the methylome (1). This theory provides the knowledge on the family of probability distributions that better fit the methylation signals when expressed in terms of information divergences of methylation levels. Because the biological signal created within the dynamic methylome environment characteristic of organisms is not free from background noise, the approach, designated Methyl-IT, includes an application of signal detection theory.
A basic requirement for the application of signal detection is the knowledge of the background noise probability distribution. A generalized gamma (GG) probability distribution model can be deduced on a statistical mechanical/thermodynamics basis for DNA methylation induced by thermal fluctuations (1), which leads to the particular cases of members of GG probability distribution family. For example, assuming that this background methylation variation is consistent with a Poisson process, it can be distinguished from variation associated with methylation regulatory machinery, which is non-independent for all genomic regions (1). An information-theoretic divergence to express the variation in methylation induced by background thermal fluctuations will follow a probability distribution model, member of the GG family, provided that it is proportional to minimum energy dissipated per bit of information from methylation change. The information thermodynamics model was previously verified with more than 150 Arabidopsis and more than 90 human methylome datasets (3).
With Methyl-IT R package we are providing the functions from the R scripts used in the manuscript (1), as well as, additional functions that will be used in further studies. The application of the information thermodynamics of cytosine DNA methylation is not limited to the current methylome analysis, which is only a particular application. The theory permits us the study of plant and animal methylomes in the framework of a communication system (3), where cytosine DNA methylation has the dual roles of stabilizing the DNA molecule and to carry the regulatory signals.
The application of Methyl-T signal detection-machine learning approach to methylation analysis of whole genome bisulfite sequencing (WGBS) data permits a high level of methylation signal resolution in cancer-associated genes and pathways (4).
Currently, the package is actively used in methylation analyses. Nevertheless, improvements are regularly introduced. Watch this repo or check for updates. THE PACKAGE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED.
Be sure that both the R and bioconductor packages are up to date. To get the latest version of Bioconductor by starting R and entering the commands:
if (!requireNamespace("BiocManager")) install.packages("BiocManager")
BiocManager::install()
Install OS packages:
CentOS 7
sudo yum -y install libcurl-devel.x86_64 openssl-devel.x86_64 libxml2-devel mariadb-devel.x86_64
Ubuntu
sudo apt install libmysqlclient-dev libcurl4-openssl-dev libssl-dev libxml2-dev git build-essential
Install R dependencies:
install.packages(c("ArgumentCheck", "caret", "Epi", "e1071", "minpack.lm", "nls2", "caTools", "rmarkdown", "RCurl"),dependencies=TRUE)
BiocManager::install(c('BiocParallel', 'biovizBase', 'genefilter', 'GenomeInfoDb', 'GenomicRanges'))
You can install MethylIT from GitHub or PSU's GitLab:
install.packages("devtools")
# Master stable version (0.3.2)
devtools::install_git("https://github.com/genomaths/MethylIT.git")
# Beta (developing) version (0.3.2_beta):
devtools::install_git("https://github.com/genomaths/MethylIT.git", ref = "MethylIT_0.3.2_beta")
Or (0.3.1)
devtools::install_git("https://git.psu.edu/genomath/MethylIT.git")
An example with simulated datasets to illustrate why a signal detection step is needed in methylation analysis.
Optimal cutpoint for the methylation signal.
Principal Components and Linear Discriminant Analyses with Methyl-IT
https://git.psu.edu/genomath/MethylIT_examples
R script and data for testing after installation
1. Sanchez, R.; Yang, X.; Maher, T.; Mackenzie, S.A. Discrimination of DNA Methylation Signal from Background Variation for Clinical Diagnostics. Int. J. Mol. Sci. 2019, 20, 5343.
2. Sanchez R, Mackenzie SA: Genome-Wide Discriminatory Information Patterns of Cytosine DNA Methylation. Int. J. Mol. Sci. 2016, 17(6), 938.
3. Sanchez R, Mackenzie SA: Information Thermodynamics of Cytosine DNA Methylation. Plos One 2016, 11.
4. Sanchez R, Mackenzie SA: Integrative Network Analysis of Differentially Methylated and Expressed Genes for Biomarker Identification in Leukemia. Sci Rep 2020, 10(1), 2123.
You are free to copy, distribute and transmit MethylIT for non-commercial purposes. Any use of MethylIT for a commercial purpose is subject to and requires a special license.
For questions about the MethylIT project, contact rus547@psu.edu
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.