The package repository is located at
https://github.com/bottom-bracket/manipylate.
It can also be installed from pypi with pip install manipylate.
In research we spend some time with plotting data, comparing measurement results
or testing the influence of parameter changes. Often the most intuitive approach
is by looking at different plots.
However, while python is great to create reproducible figures, I found no way to
quickly create plots with which one can interact without changing the code used
to create the plot as in mathematica’s Manipulate. The closest one can get
to this is interact from ipywidgets which is limited to run in Jupyter
notebooks.
Manipylate shall provide the simplified functionalities from its bigger sibling Manipulate. The tasks I had in mind while writing the tool are:
- Plotting 1d and 2d cuts of multidimensional data and changing the slice index with sliders
- Plot a function for changing parameters which one can modify interactively
I also wanted to keep things as simple as possible and be able to use
manipylate outside jupyter notebooks.
- Sliders are added automatically
- Views can be saved with
Shift + middle mouse clickin the figure if used in purematplotlibmode and by click on the camera button injupyter
First we get the environment ready (leave out the lines starting with % for
non-ipython shells).
%matplotlib qt
%load_ext autoreload
%autoreload 2
import matplotlib as mpl
from matplotlib import pyplot as plt
import numpy as np
from manipylate import ifigure
Let’s create some data:
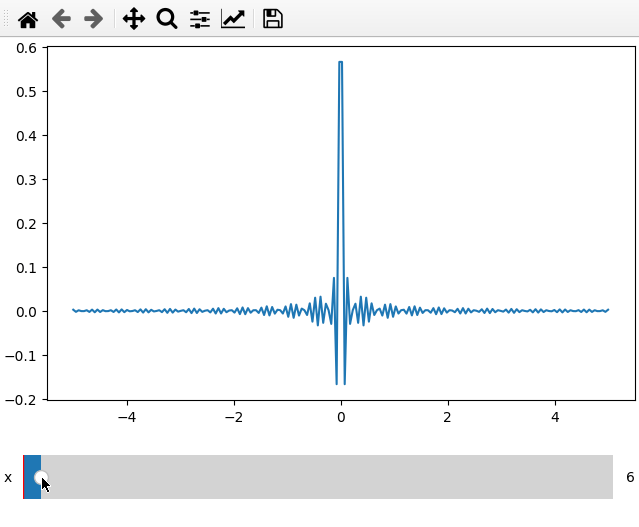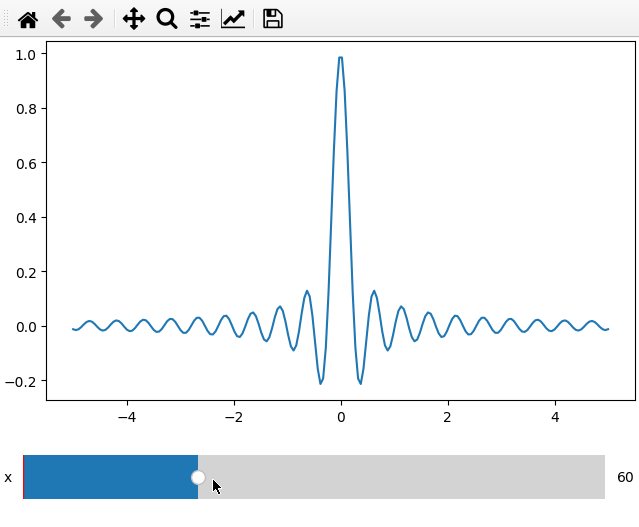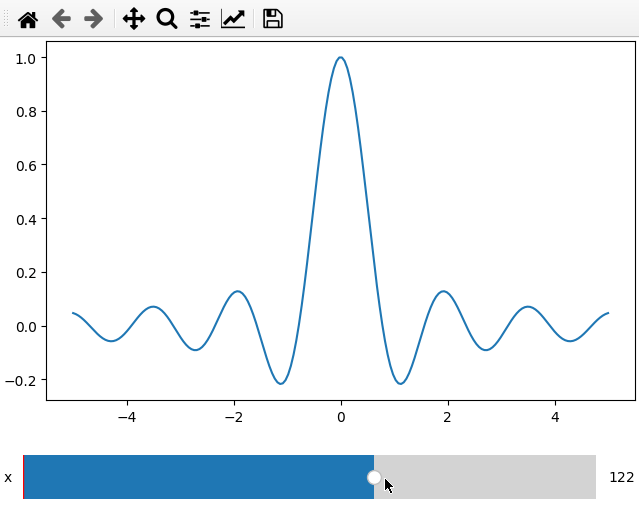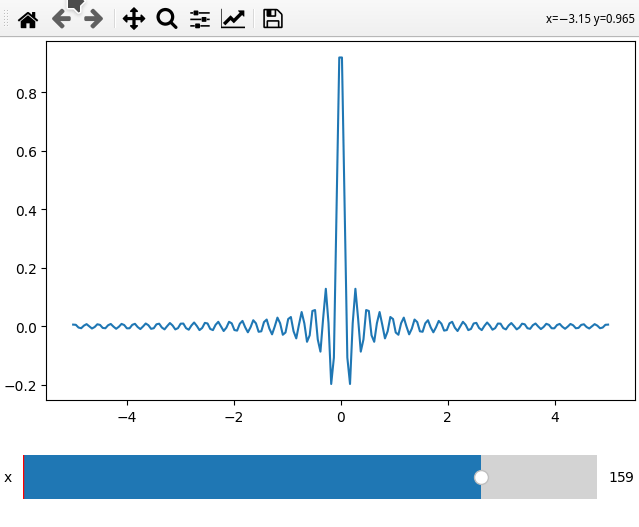
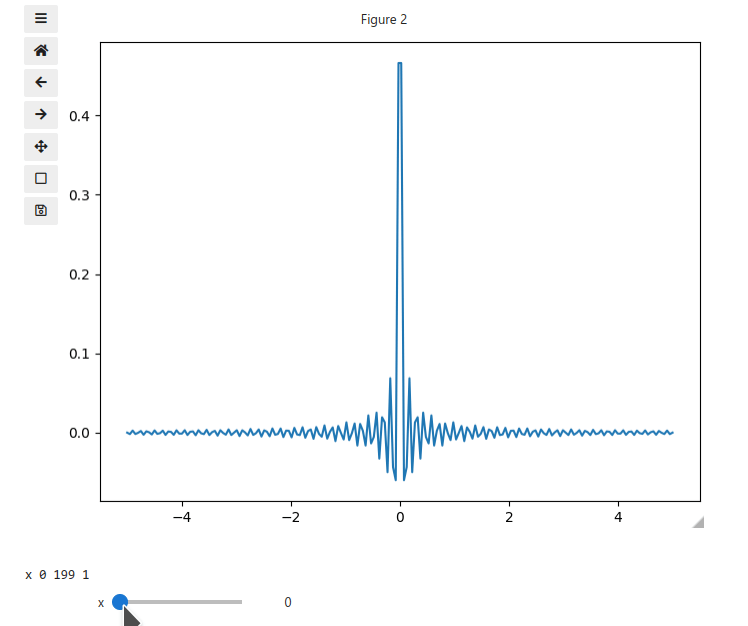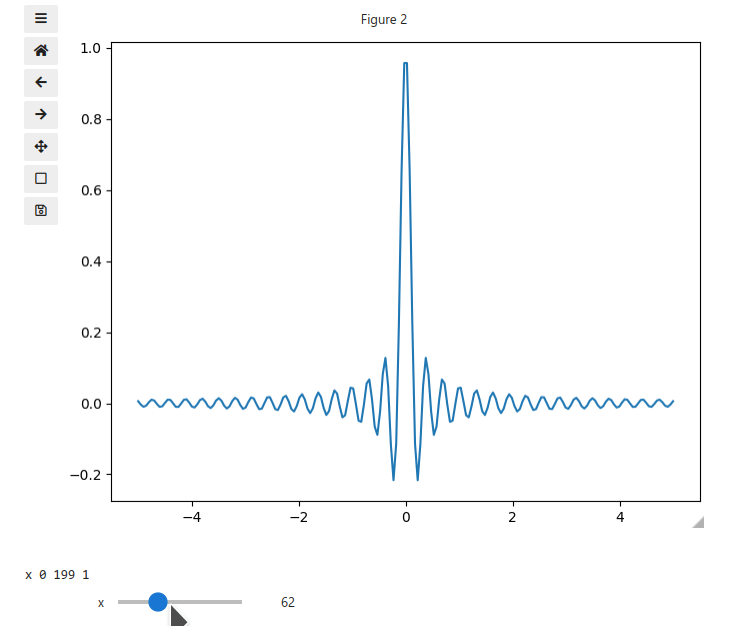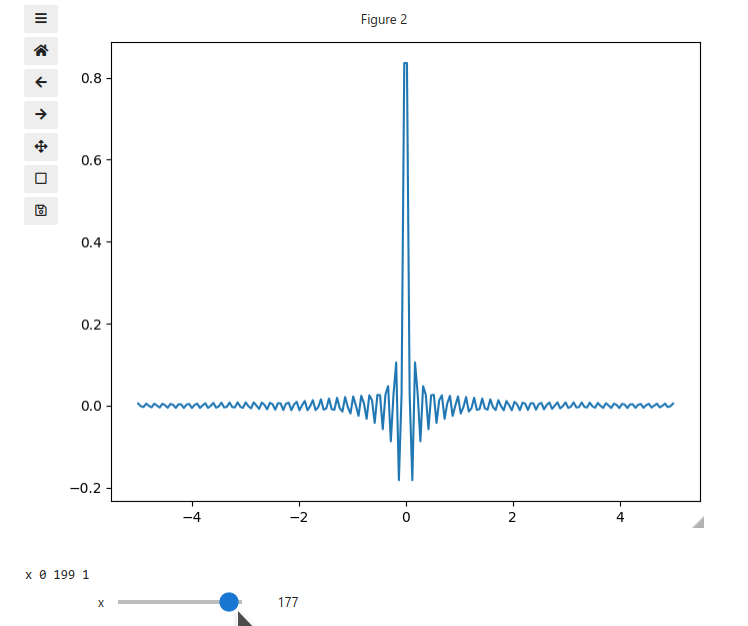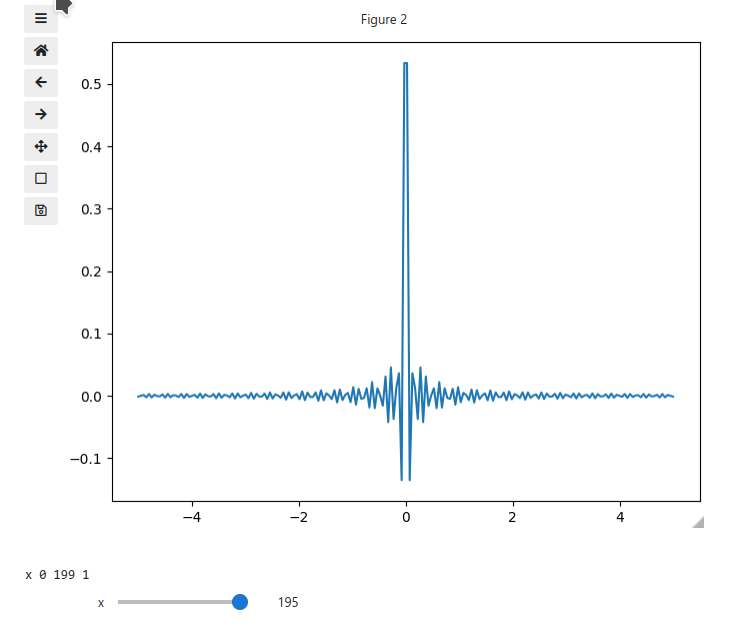
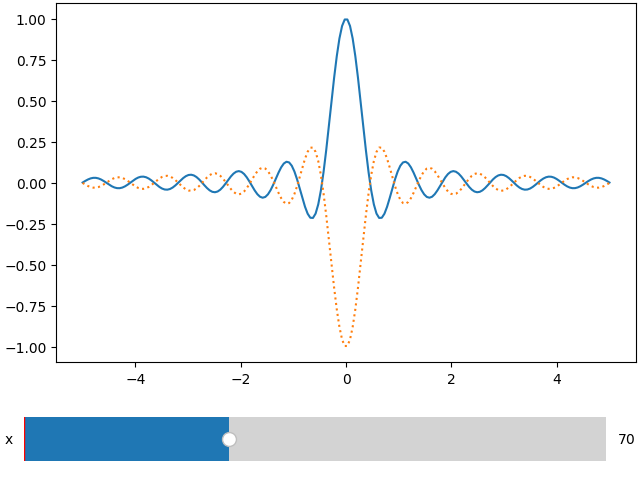
x = np.linspace(-5, 5, 200)
X, Y = np.meshgrid(x, x)
data = np.sinc(X * Y**2)
The figure is created by creating an ifigure object. We can then call its
add_plot method to add an axis and a line plot. Here axpos defines where the
plot shall be placed, x is the x-axis data. data the 2d array of which we
want to plot a cut along the axis given by plot_ax, and parameters is a list
of dimension parameters we want to change using sliders.
Calling show creates the figure. It needs to be called after all plots have been
added to the figure since here we’ll create the sliders and arange the slider
axes (in case of execution outside jupyter).
ifig = ifigure(1,1)
ifig.add_plot(axpos=[0,0],x=x,data=data,parameters=['x'],plot_ax=1)
ifig.show()
In order to use the ipywidget widgets, we call ifigure with the
jupyterwidget keyword argument.
ifig = ifigure(1,1,jupyterwidget=True)
ifig.add_plot(axpos=[0,0],x=x,data=data,parameters=['x'],plot_ax=1)
ifig.show()
Axes are remembered by the ifigure object based on their position, so we can
call the same add_plot command with another dataset. Note that any keyword
arguments are passed to matplotlib.axes.plot, so we can use this to change the
line style.
ifig = ifigure(1,1)
ifig.add_plot(axpos=[0,0],x=x,data=data,parameters=['x'],plot_ax=1)
ifig.add_plot(axpos=[0,0],x=x,data=-data,parameters=['x'],plot_ax=1,ls=':')
ifig.show()
Note that here both plots share the same parameter. In the next example we will see that if called with a different name, a new slider will be created.
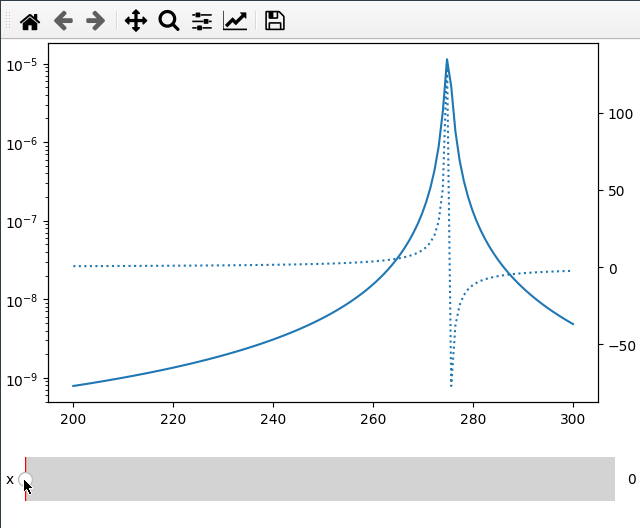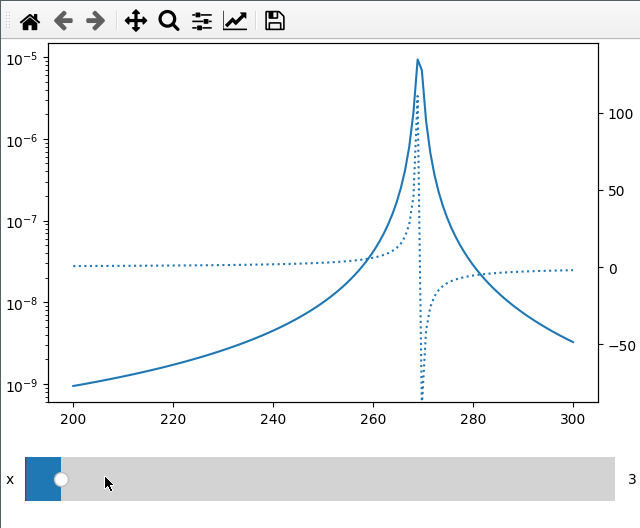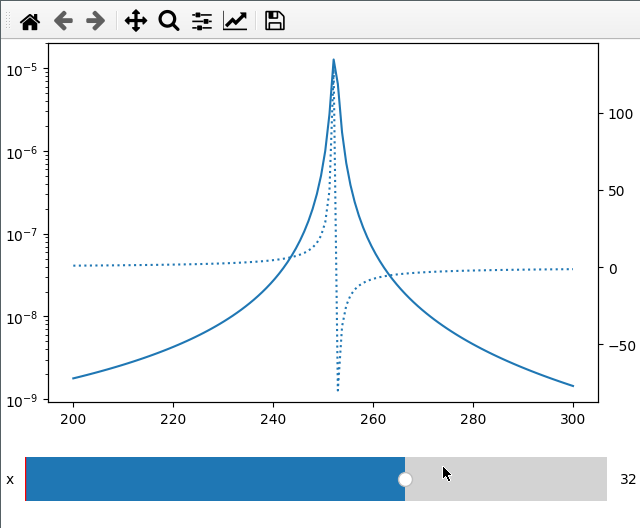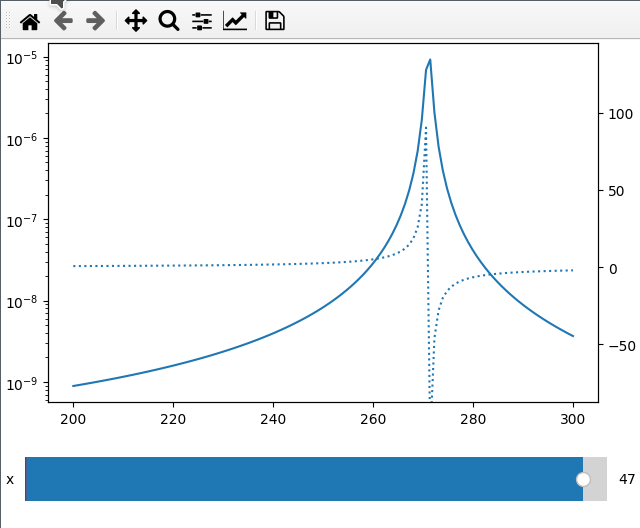
Sometimes I want to compare two different values (like amplitude and phase) over
the same x axis. This can be done by passing the twinx=True to the add_plot
method. Here I also use the fix_lim flag to prevent the phase axis from
updating.
Note that add_plot returns a matplotlib axis instance which we can modify as
usual (here setting the y scale to logarithmic).
fc = np.linspace(-5, 5, 50) ** 2 + 250
x = np.linspace(200, 300, 120)
FC, X = np.meshgrid(fc, x, indexing="ij")
data = 1 / ((FC ** 2 - X ** 2 - 1j * X * 1) ** 2)
ifig = ifigure(1, 1)
ax = ifig.add_plot(axpos=[0, 0], x=x, data=np.abs(data), parameters=["fc"], plot_ax=1)
ax.set_yscale("log")
ifig.add_plot(
axpos=[0, 0],
x=x,
data=np.angle(data, deg=True),
parameters=["x"],
plot_ax=1,
twinx=True,
ls=":",
fix_lim=True,
)
ifig.show()
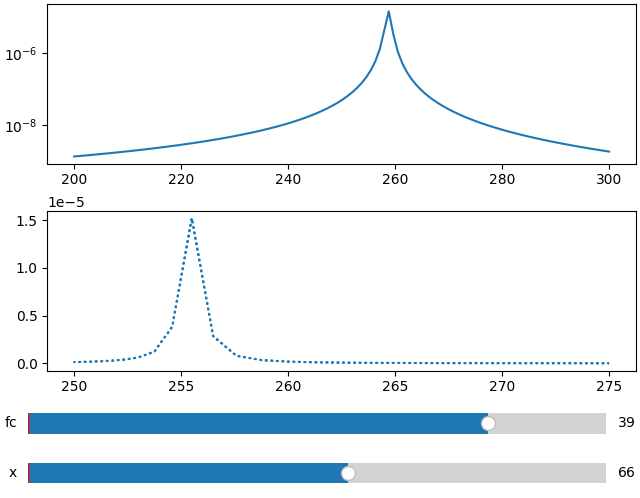
In order to add more than one subplots we adjust the ifigure creation.
Here we also use another parameter for the second plot.
fc = np.linspace(-5, 5, 50) ** 2 + 250
x = np.linspace(200, 300, 120)
FC, X = np.meshgrid(fc, x, indexing="ij")
data = 1 / ((FC ** 2 - X ** 2 - 1j * X * 1) ** 2)
ifig = ifigure(2, 1)
ax = ifig.add_plot(axpos=[0, 0], x=x, data=np.abs(data), parameters=["fc"], plot_ax=1)
ax.set_yscale("log")
ifig.add_plot(
axpos=[1, 0],
x=fc,
data=np.abs(data),
parameters=["x"],
plot_ax=0,
ls=":",
)
ifig.show()
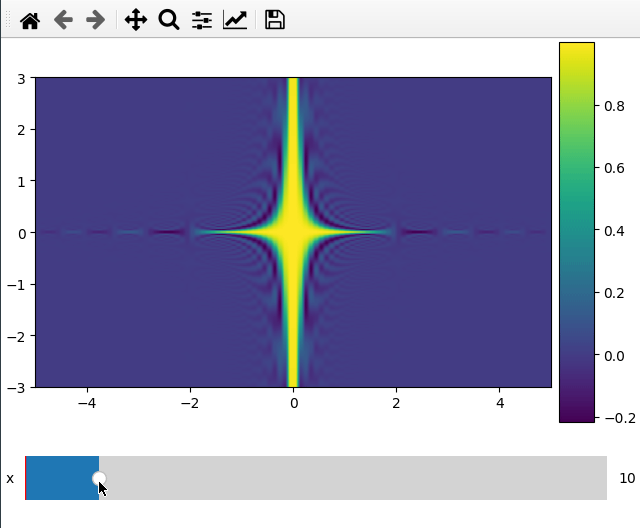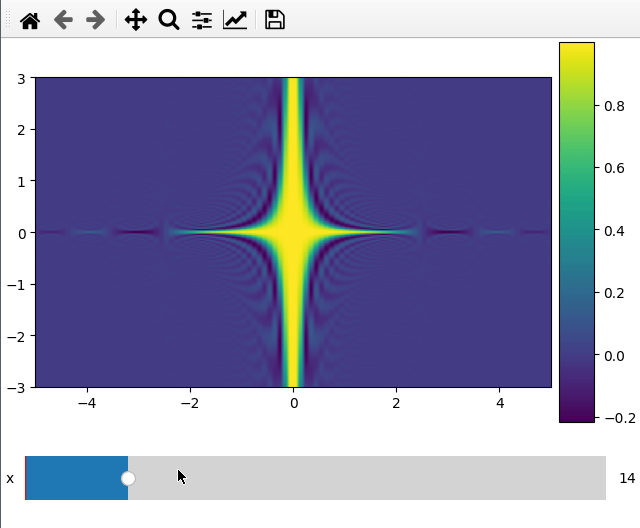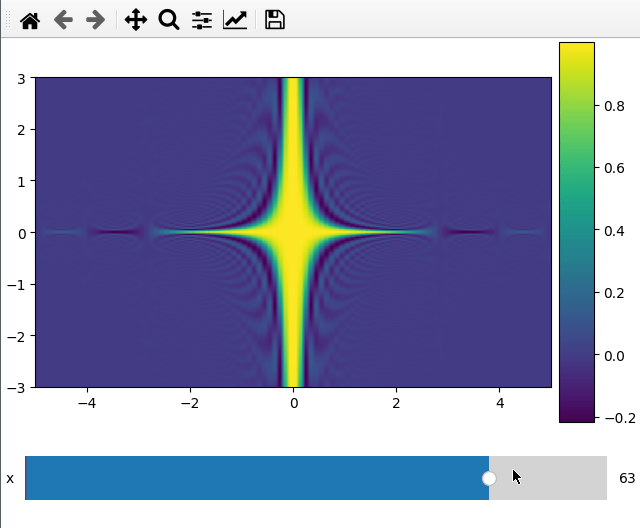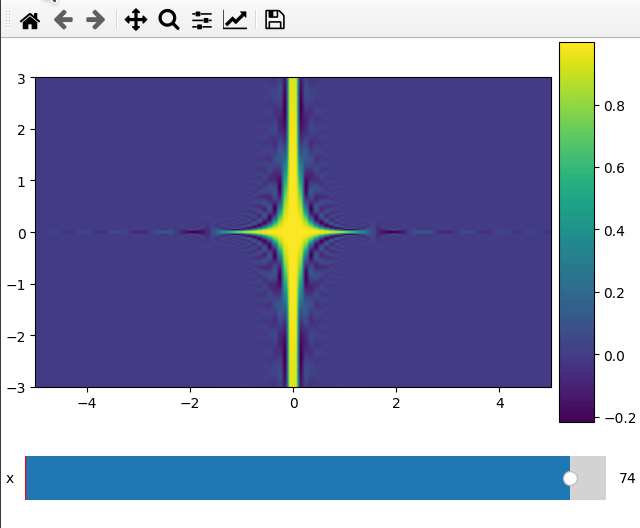
Plotting a 2d cut is nearly the same as a 1d line. ifigure creates an imshow
plot when being passed data which has two more dimensions than the number of
parameters passed to the function.
For the 2d plot we need to specify along which axes we want to cut by changing
the plot_ax argument to a list of length 2.
x = np.linspace(-5, 5, 200)
y = np.linspace(-3, 3, 100)
z = np.linspace(-4, 4, 80)
X, Y ,Z = np.meshgrid(x, y, z,indexing='ij')
data = np.sinc(X * Y**2 * Z**3)
ifig=ifigure(1,1)
ifig.add_plot(axpos=[0,0],x=[x,y],data=data,parameters=['x'],plot_ax=[0,1])
ifig.show()
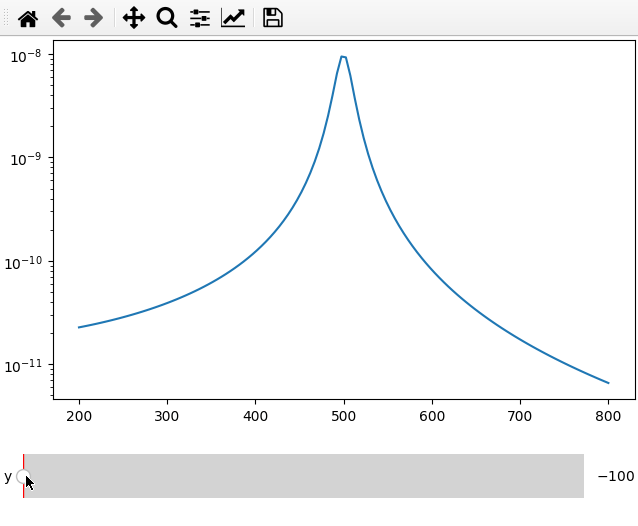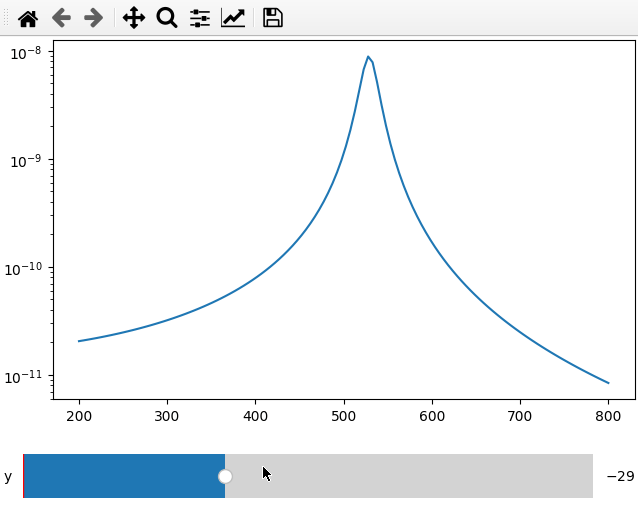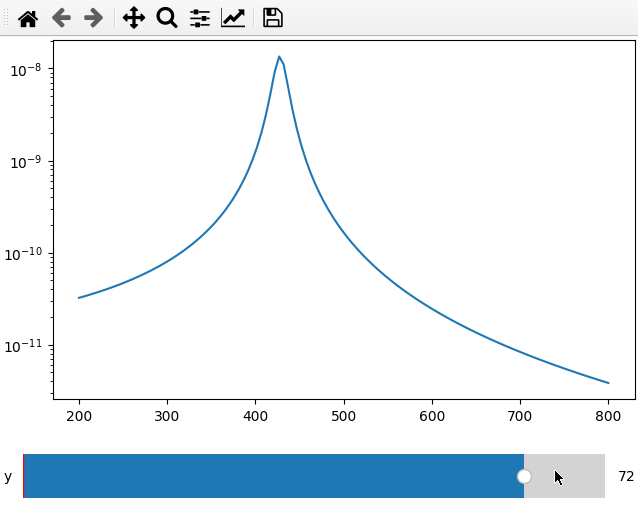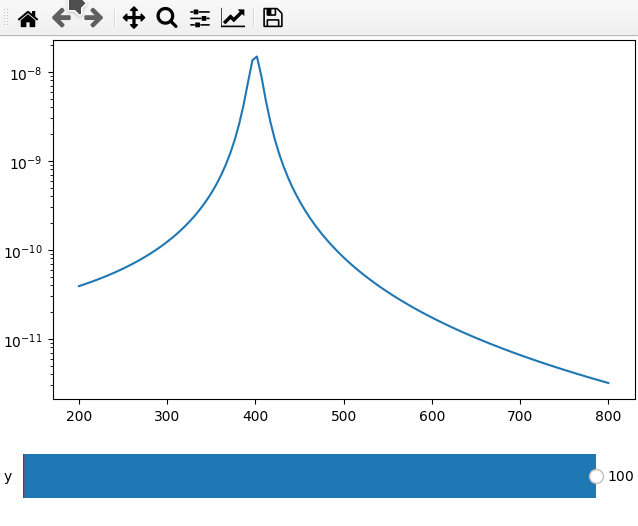
The data argument can be replaced by a function that returns either a 1d
array for a line plot or a 2d array for a map plot.
The main difference to calling the plot on an array is that we do not need to
specify the plot_ax parameter but we need to define a range and step size for
the slider, which is done by replacing the string argument in the parameter list
by a list containing name,minimum, maximum and step size.
fc = 250
x = np.linspace(200, 800, 120)
def lor(y):
return np.abs(1 / ((4*fc ** 2 - (x+y) ** 2 - 1j * x * 20) ** 2))
ifig = ifigure(1, 1)
ax = ifig.add_plot(axpos=[0, 0], x=x, data=lor, parameters=[["y",-100,100,1]])
ax.set_yscale("log")
ifig.show()
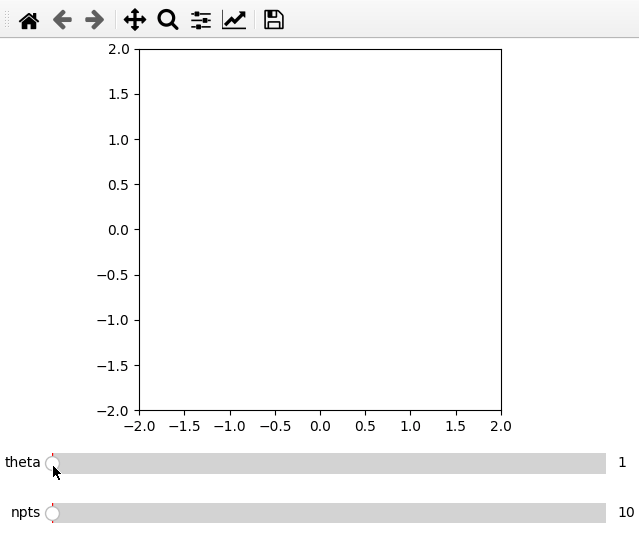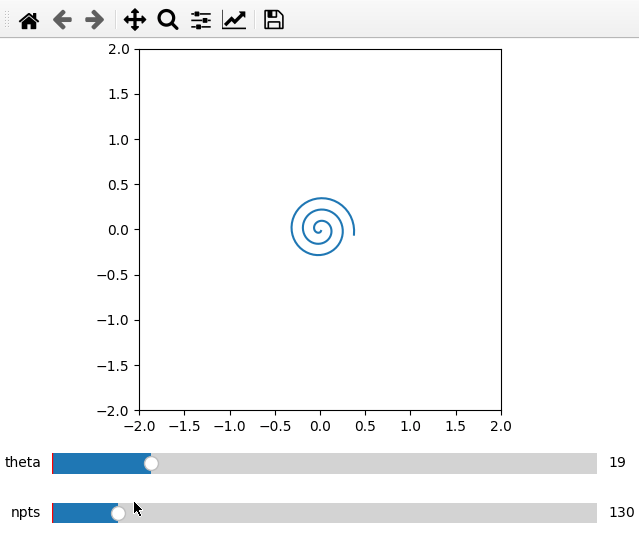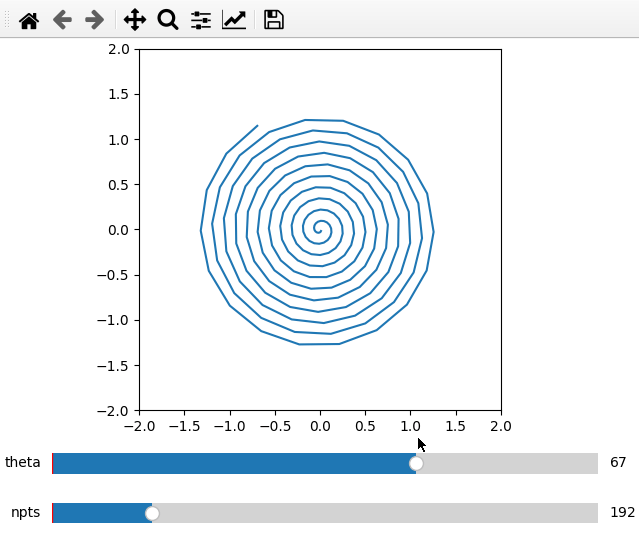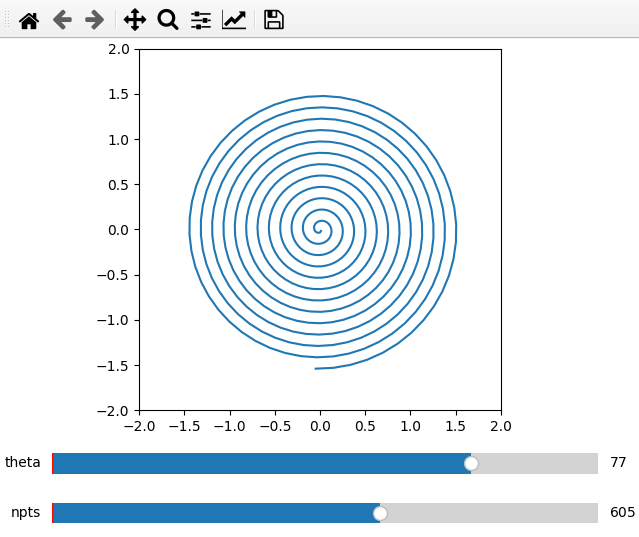
Furthermore it is possible to do parametric plots using a function for x and
data arguments. Make sure that the variable names are the same in the
definition and add_plot call.
import numpy as np
from manipylate import ifigure
def x(theta,npts):
th = np.linspace(1,theta,npts)
return th/50*np.cos(th)
def y(theta,npts):
th = np.linspace(1,theta,npts)
return -th/50*np.sin(th)
ifig = ifigure(1,1)
ax = ifig.add_plot(axpos=[0,0],x=x,data=y,parameters=[['theta',1,100,1],['npts',10,1000,1]],fix_lim=True)
ax.set_ylim(-2,2)
ax.set_xlim(-2,2)
ax.set_aspect('equal')
ifig.show()
If one of the functions x,y takes more arguments than the other just add a
**kwargs to its definition (e.g. def x(theta,npts,**kwargs)).
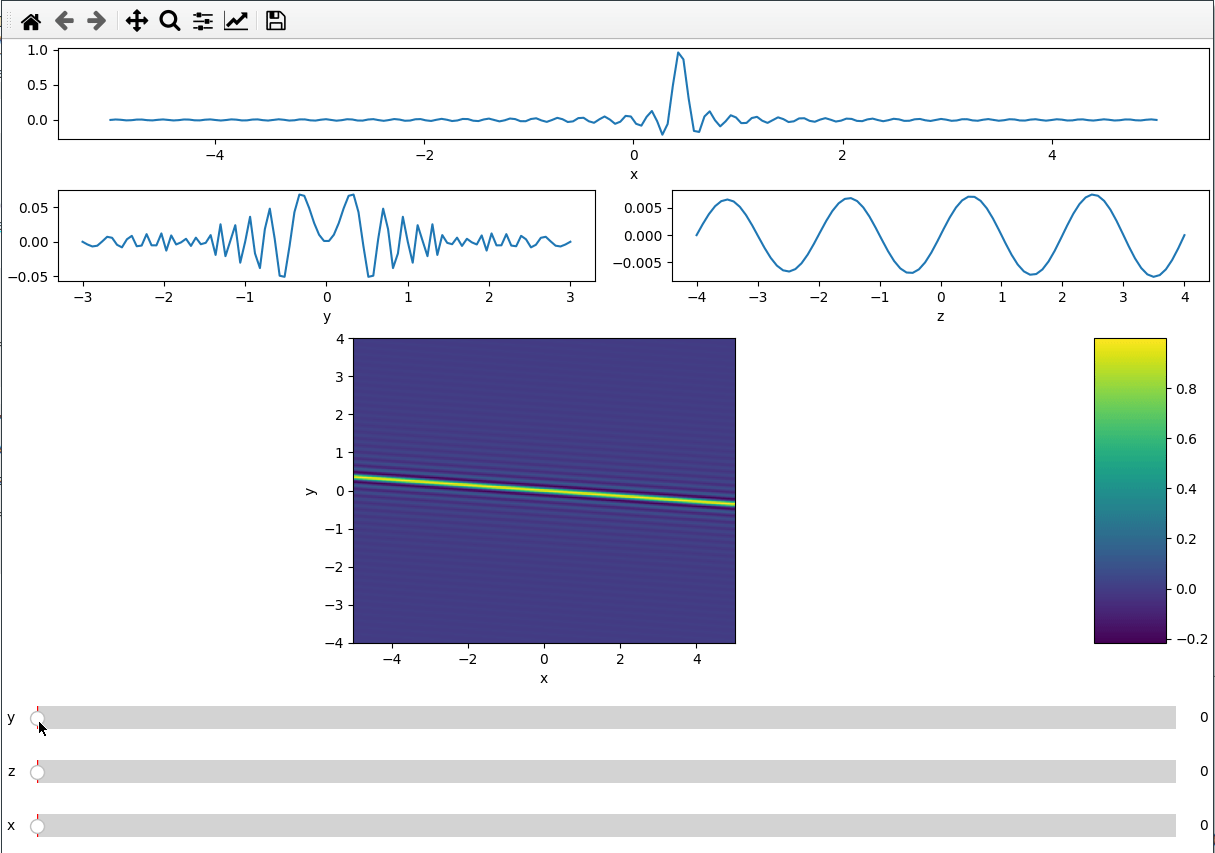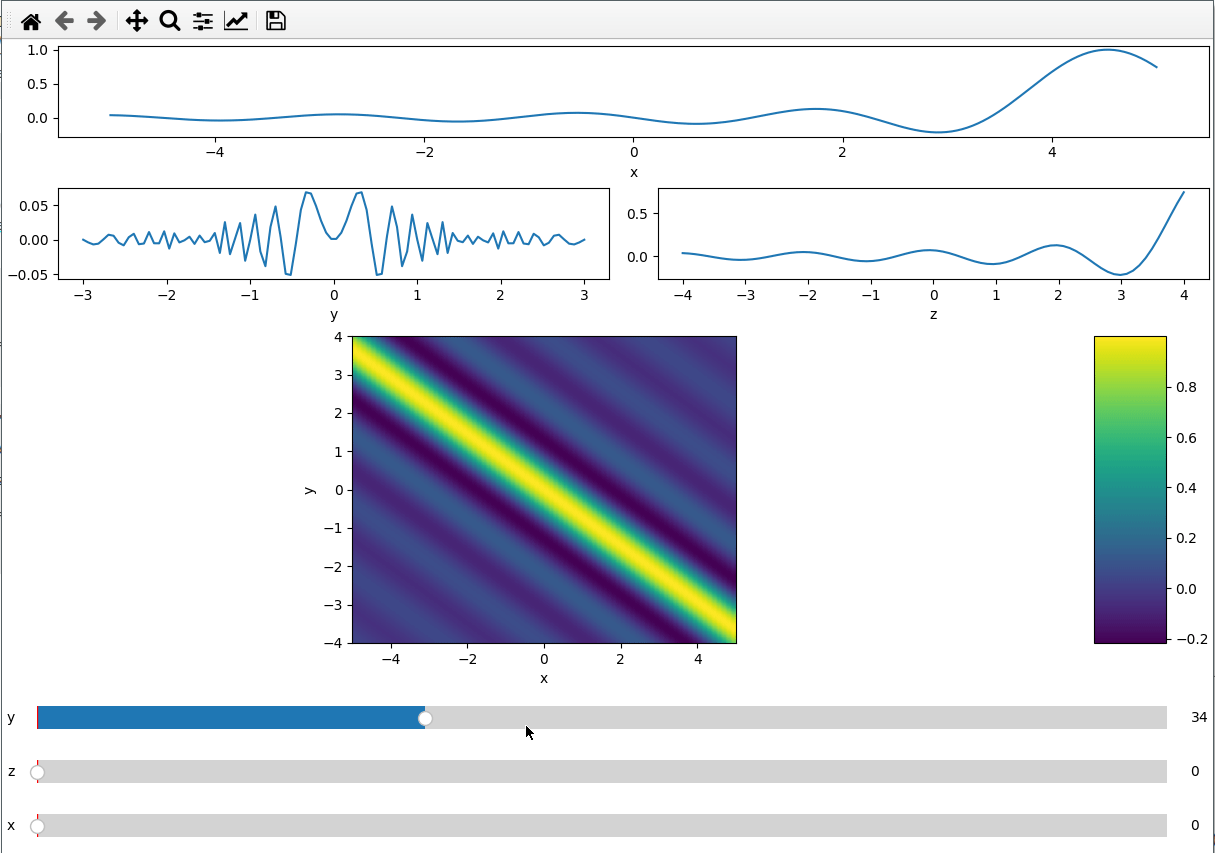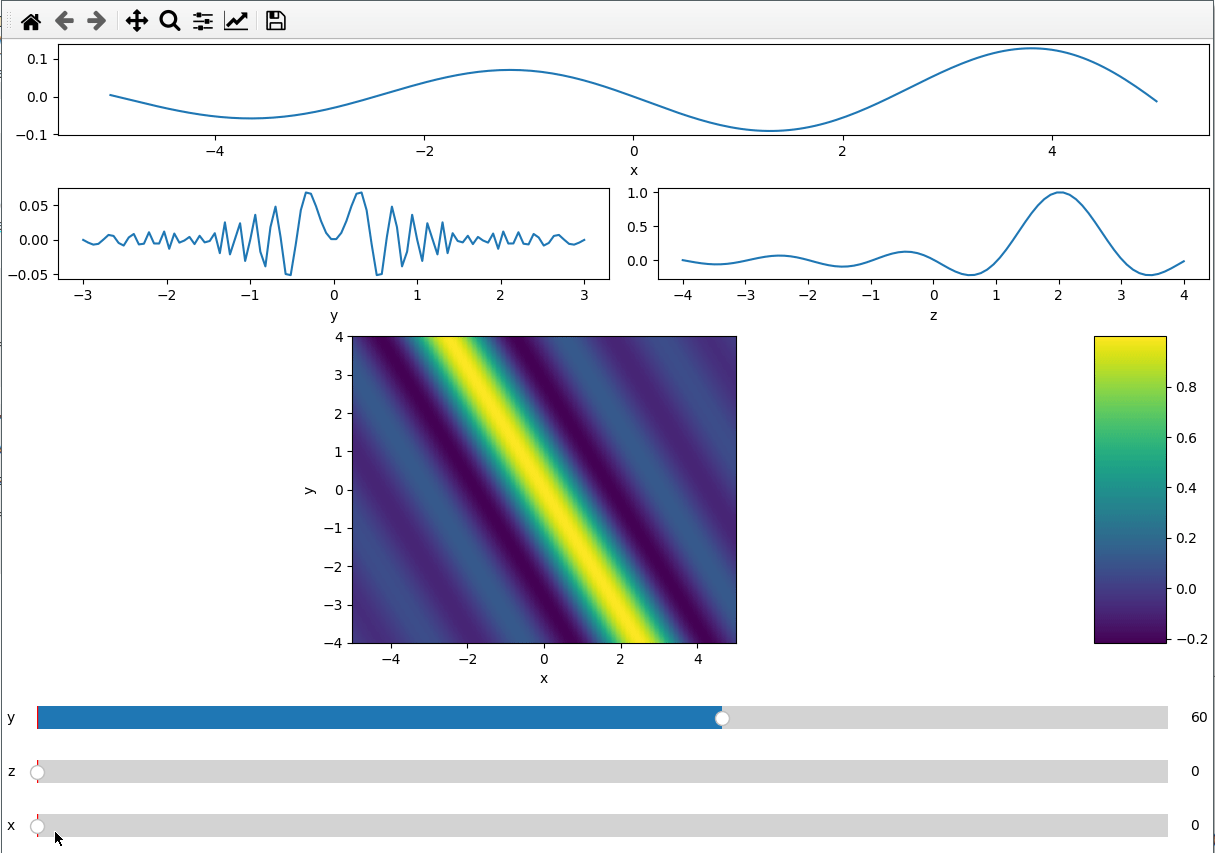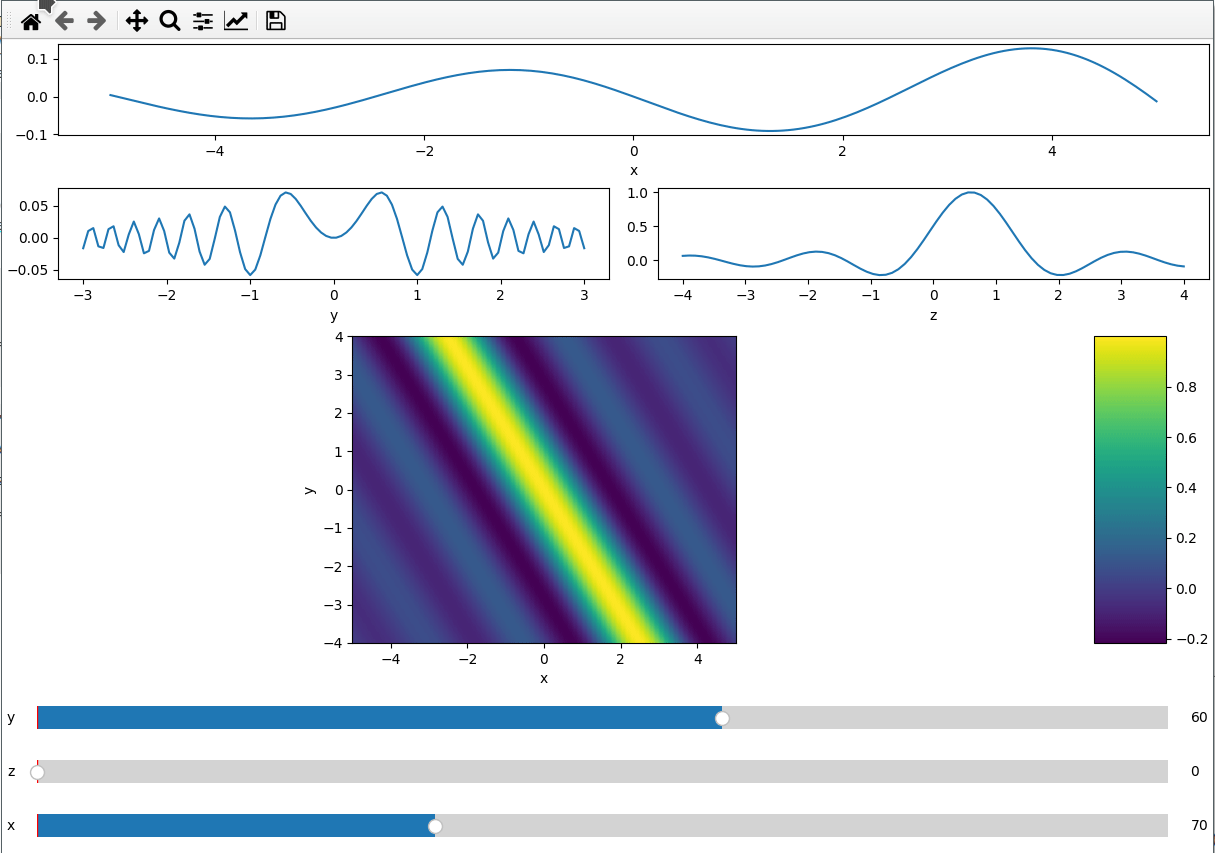
Since the subplot layout creation is based on GridSpec, we can create more
complicated layouts. We use the handy numpy.s_ to create the exact slices to
index the GridSpec.
x = np.linspace(-5, 5, 200)
y = np.linspace(-3, 3, 100)
z = np.linspace(-4, 4, 80)
X, Y ,Z = np.meshgrid(x, y, z,indexing='ij')
data = np.sinc(X * Y**2 + Z)
ifig = ifigure(6, 4,figsize=(12,8))
ax=ifig.add_plot(axpos=np.s_[0,:], x=x, data=data, parameters=["y",'z'],plot_ax=0)
ax.set(xlabel='x')
ax=ifig.add_plot(axpos=np.s_[1,:2], x=y, data=data, parameters=["x","z"],plot_ax=1)
ax.set(xlabel='y')
ax=ifig.add_plot(axpos=np.s_[1,2:], x=z, data=data, parameters=["x","y"],plot_ax=2)
ax.set(xlabel='z')
ax=ifig.add_plot(axpos=np.s_[2::,:], x=[x,z], data=data, parameters=["y"],plot_ax=[0,2])
ax.set(xlabel='x',ylabel='y')
ifig.show()
The ifigure class can be called with the figurename parameter which will
determine the base name and the extension for individually saved views. The full
filename is constructed from the figurename and the current slider names and
values.
Slider axes are removed during saving.
- Comment and document code
- add
convert_parammethod to display physical parameter values (e.g. 0-1μm instead of index values 1-51) - 3d plots ?
- choice for 2d plots (contour)