Histo-Miner: Tissue Features Extraction With Deep Learning from H&E Images of Squameous Cell Carcinoma Skin Cancer
The README will be comprehensive when the corresponding paper presenting histo-miner will be available on arxiv. So far the README is not complete.
Histo-Miner presentation • Histo-Miner visualization • Installation • Project Structure • Usage • Examples • Roadmap
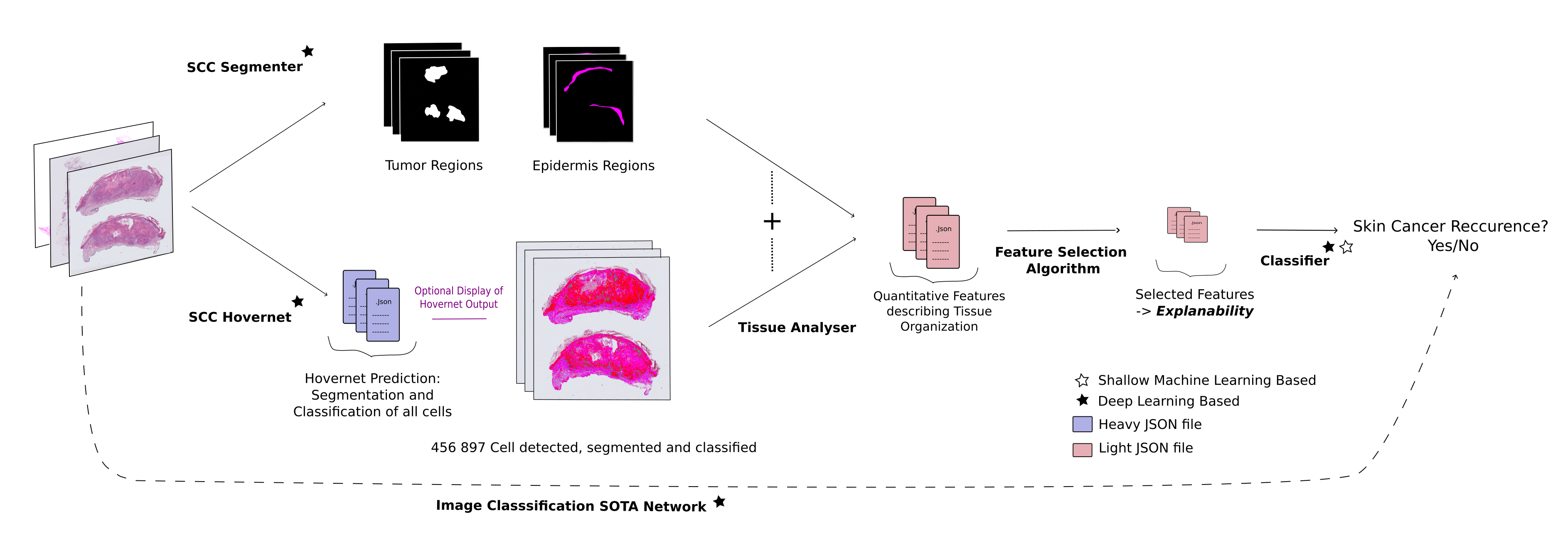
Histo-Miner employs convolutional neural networks and vision transformers models for nucleus segmentation and classification as well as tumor region segmentation. From these predictions, it generates a compact feature vector summarizing tissue morphology and cellular interactions. We used such generated features to classify cSCC patient response to immunotherapy.
Here is an explanation of the project structure:
├── checkpoints # README defining where to find our models checkpoints in Zenodo
├── configs # All configs file with explanations
│ ├── models # example configs for both models inference
│ ├── classification_training # Configs for classifier training
│ ├── histo_miner_pipeline # Configs for the core code of histo-minerent
├── datasets # README defining where to find our datasets in Zenodo
├── docs # Documentation files (in addition to this main README.md)
├── scripts # Main code for users to run Histo-Miner
├── src # Functions used for scripts
│ ├── histo-miner # All functions from the core code (everything except deep learning)
│ ├── models # Submodules of models for inference and training
│ │ ├── hover-net # hover net submodule, simplification of original code to fit histo-miner needs
│ │ ├── mmsegmentation # segmenter submodule, simplification of original code to fit histo-miner needs
├── vizualization # Both python and groovy scripts to either reproduce paper figures or to vizualize model inference with qupath