This remote module of the Orfeo ToolBox provides a generic, multi purpose deep learning framework, targeting remote sensing images processing. It contains a set of new process objects that internally invoke Tensorflow, and a bunch of user-oriented applications to perform deep learning with real-world remote sensing images. Applications can be used to build OTB pipelines from Python or C++ APIs.
- Sampling,
- Training, supporting save/restore/import operations (a model can be trained from scratch or fine-tuned),
- Serving models with support of OTB streaming mechanism. Meaning (1) not limited by images sizes, (2) can be used as a "lego" in any OTB pipeline and preserve streaming, (3) MPI support available (use multiple processing unit to generate one single output image)
Below are some screen captures of deep learning applications performed at large scale with OTBTF.
- Image to image translation (Spot-7 image --> Wikimedia Map using CGAN)
- Landcover mapping (Spot-7 images --> Building map using semantic segmentation)
- Image enhancement (Enhancement of Sentinel-2 images at 1.5m using SRGAN)
You can read more details about these applications on this blog
For now you have two options: either use the existing docker image, or build everything yourself from source.
Use the latest image from dockerhub:
docker pull mdl4eo/otbtf2.0:cpu
docker run -u otbuser -v $(pwd):/home/otbuser mdl4eo/otbtf2.0:cpu otbcli_PatchesExtraction -help
Available docker images:
| Name | Os | TF | OTB | Description |
|---|---|---|---|---|
| mdl4eo/otbtf1.6:cpu | Ubuntu Xenial | r1.14 | 7.0.0 | CPU, no optimization |
| mdl4eo/otbtf1.7:cpu | Ubuntu Xenial | r1.14 | 7.0.0 | CPU, no optimization |
| mdl4eo/otbtf1.7:gpu | Ubuntu Xenial | r1.14 | 7.0.0 | GPU |
| mdl4eo/otbtf2.0:cpu | Ubuntu Xenial | r2.1 | 7.1.0 | CPU, compiled with MKL |
| mdl4eo/otbtf2.0:gpu | Ubuntu Xenial | r2.1 | 7.1.0 | GPU |
All GPU docker images are suited for NVIDIA GPUs. They use CUDA/CUDNN support and are built with compute capabilities 6.1, 5.2, 3.5. To change the compute capabilities, you can edit the dockerfile and build your own docker image (dockerfiles are provided in the tools/dockerfiles/ path of the repository).
You can find more details on the GPU docker image and some docker tips and tricks on this blog
See here to see how to build the remote module from sources.
Let's describe quickly the new applications provided.
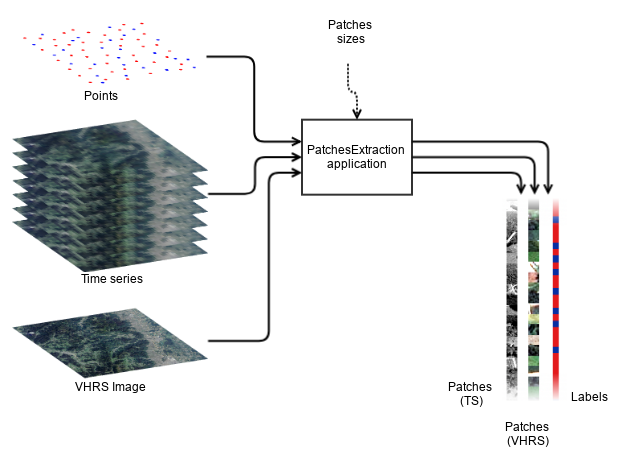
This application performs the extraction of patches in images from a vector data containing points.
The OTB sampling framework can be used to generate the set of selected points.
After that, you can use the PatchesExtraction application to perform the sampling of your images.
We denote input source an input image, or a stack of input images (of the same size !).
The user can set the OTB_TF_NSOURCES environment variable to select the number of input sources that he wants.
For example, for sampling a Time Series (TS) together with a single Very High Resolution image (VHR), a number of 2 sources is required: 1 input images list for time series and 1 input image for the VHR.
The sampled patches will be extracted at each positions designed by the points, only if they are entirely lying inside all input sources extents.
For each input source, patches sizes must be provided.
For each input source, the application export all sampled patches as a single multiband raster, stacked in rows.
For instance, for n samples of size 16 x 16 from a 4 channels input source, the output image will be a raster of size 16 x 16n with 4 channels.
An optional output is an image of size 1 x n containing the value of one specific field of the input vector data.
Typically, the class field can be used to generate a dataset suitable for a model that performs pixel wise classification.
This application extracts patches in multiple input images. Change the OTB_TF_NSOURCES environment variable to set the number of sources.
Parameters:
-source1 <group> Parameters for source 1
MISSING -source1.il <string list> Input image(s) 1 (mandatory)
MISSING -source1.out <string> [pixel] Output patches for image 1 [pixel=uint8/uint16/int16/uint32/int32/float/double/cint16/cint32/cfloat/cdouble] (default value is float) (mandatory)
MISSING -source1.patchsizex <int32> X patch size for image 1 (mandatory)
MISSING -source1.patchsizey <int32> Y patch size for image 1 (mandatory)
-source1.nodata <float> No-data value for image 1(used only if "usenodata" is on) (mandatory, default value is 0)
MISSING -vec <string> Positions of the samples (must be in the same projection as input image) (mandatory)
-usenodata <boolean> Reject samples that have no-data value (optional, off by default, default value is false)
-outlabels <string> [pixel] output labels [pixel=uint8/uint16/int16/uint32/int32/float/double/cint16/cint32/cfloat/cdouble] (default value is uint8) (optional, off by default)
MISSING -field <string> field of class in the vector data (mandatory)
-progress <boolean> Report progress
-help <string list> Display long help (empty list), or help for given parameters keys
Use -help param1 [... paramN] to see detailed documentation of those parameters.
Examples:
otbcli_PatchesExtraction -vec points.sqlite -source1.il $s2_list -source1.patchsizex 16 -source1.patchsizey 16 -field class -source1.out outpatches_16x16.tif -outlabels outlabels.tif
You can build models using the TensorFlow Python API as shown in the ./python/ directory.
Models must be exported in SavedModel format.
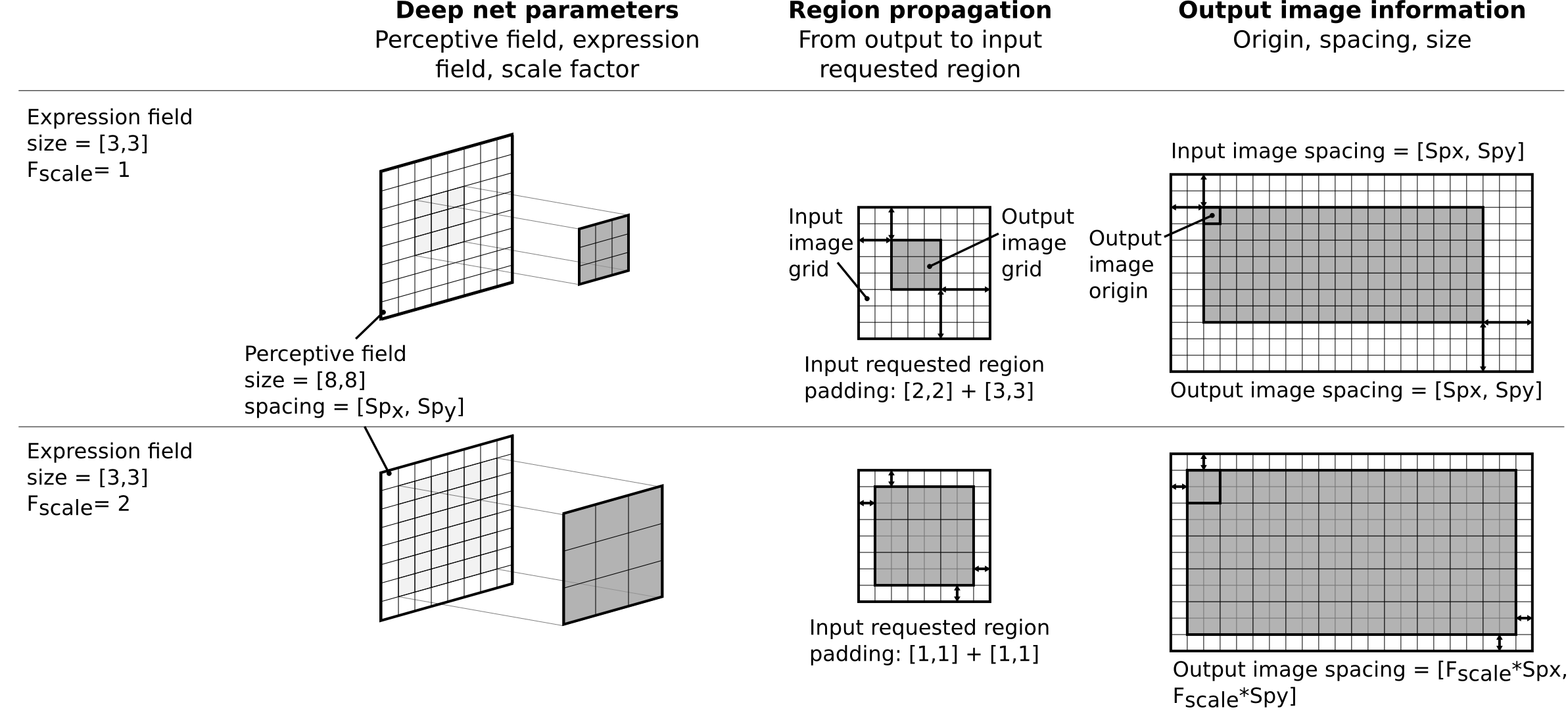
When using a model in OTBTF, the important thing is to know the following parameters related to the placeholders (the inputs of your model) and output tensors (the outputs of your model).
- For each input placeholder:
- Name
- Receptive field
- For each output tensor:
- Name
- Expression field
- Scale factor
The scale factor descibes the physical change of spacing of the outputs, typically introduced in the model by non unitary strides in pooling or convolution operators. For each output, it is expressed relatively to one single input of the model called the reference input source. Additionally, the names of the target nodes must be known (e.g. "optimizer"). Also, the names of user placeholders, typically scalars placeholders that are used to control some parameters of the model, must be know (e.g. "dropout_rate"). The receptive field corresponds to the input volume that "sees" the deep net. The expression field corresponds to the output volume that the deep net will create.
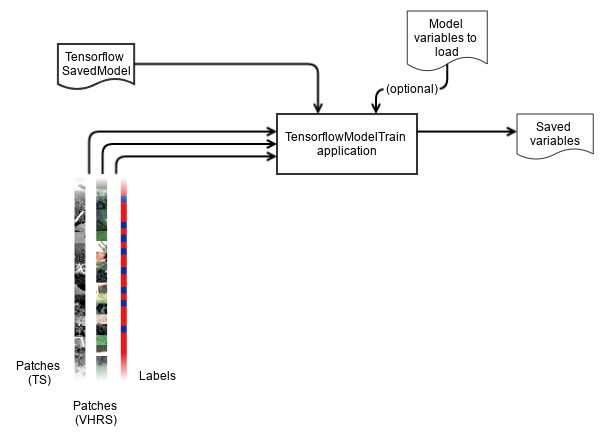
Here we assume that you have produced patches using the PatchesExtraction application, and that you have a SavedModel stored in a directory somewhere on your filesystem.
The TensorflowModelTrain application performs the training, validation (against test dataset, and against validation dataset) providing the usual metrics that machine learning frameworks provide (confusion matrix, recall, precision, f-score, ...).
You must provide the path of the SavedModel to the model.dir parameter.
The model.restorefrom and model.saveto corresponds to the variables of the SavedModel used respectively for restoring and saving them.
Set you input sources for training (training parameter group) and for validation (validation parameter group): the evaluation is performed against training data, and optionally also against the validation data (only if you set validation.mode to "class").
For each input sources, the patch size and the placeholder name must be provided.
Regarding validation, if a different name is found in a particular input source of the validation parameter group, the application knows that the input source is not fed to the model at inference, but is used as reference to compute evaluation metrics of the validation dataset.
Batch size (training.batchsize) and number of epochs (training.epochs) can be set.
User placeholders can be set separately for training (training.userplaceholders) and validation (validation.userplaceholders).
The validation.userplaceholders can be useful if you have a model that behaves differently depending the given placeholder.
Let's take the example of dropout: it's nice for training, but you have to disable it to use the model at inference time.
Hence you will pass a placeholder with "dropout_rate=0.3" for training and "dropout_rate=0.0" for validation.
Of course, one can train models from handmade python code: to import the patches images, a convenient method consist in reading patches images as numpy arrays using OTB applications (e.g. ExtractROI) or GDAL, then do a np.reshape to the dimensions wanted.
Train a multisource deep learning net using Tensorflow. Change the OTB_TF_NSOURCES environment variable to set the number of sources.
Parameters:
-model <group> Model parameters
MISSING -model.dir <string> Tensorflow model_save directory (mandatory)
-model.restorefrom <string> Restore model from path (optional, off by default)
-model.saveto <string> Save model to path (optional, off by default)
-training <group> Training parameters
-training.batchsize <int32> Batch size (mandatory, default value is 100)
-training.epochs <int32> Number of epochs (mandatory, default value is 100)
-training.userplaceholders <string list> Additional single-valued placeholders for training. Supported types: int, float, bool. (optional, off by default)
MISSING -training.targetnodes <string list> Names of the target nodes (mandatory)
-training.outputtensors <string list> Names of the output tensors to display (optional, off by default)
-training.usestreaming <boolean> Use the streaming through patches (slower but can process big dataset) (optional, off by default, default value is false)
-training.source1 <group> Parameters for source #1 (training)
MISSING -training.source1.il <string list> Input image (or list to stack) for source #1 (training) (mandatory)
MISSING -training.source1.patchsizex <int32> Patch size (x) for source #1 (mandatory)
MISSING -training.source1.patchsizey <int32> Patch size (y) for source #1 (mandatory)
MISSING -training.source1.placeholder <string> Name of the input placeholder for source #1 (training) (mandatory)
-training.source2 <group> Parameters for source #2 (training)
MISSING -training.source2.il <string list> Input image (or list to stack) for source #2 (training) (mandatory)
MISSING -training.source2.patchsizex <int32> Patch size (x) for source #2 (mandatory)
MISSING -training.source2.patchsizey <int32> Patch size (y) for source #2 (mandatory)
MISSING -training.source2.placeholder <string> Name of the input placeholder for source #2 (training) (mandatory)
-validation <group> Validation parameters
-validation.step <int32> Perform the validation every Nth epochs (mandatory, default value is 10)
-validation.mode <string> Metrics to compute [none/class/rmse] (mandatory, default value is none)
-validation.userplaceholders <string list> Additional single-valued placeholders for validation. Supported types: int, float, bool. (optional, off by default)
-validation.usestreaming <boolean> Use the streaming through patches (slower but can process big dataset) (optional, off by default, default value is false)
-validation.source1 <group> Parameters for source #1 (validation)
-validation.source1.il <string list> Input image (or list to stack) for source #1 (validation) (mandatory)
-validation.source1.name <string> Name of the input placeholder or output tensor for source #1 (validation) (mandatory)
-validation.source2 <group> Parameters for source #2 (validation)
-validation.source2.il <string list> Input image (or list to stack) for source #2 (validation) (mandatory)
-validation.source2.name <string> Name of the input placeholder or output tensor for source #2 (validation) (mandatory)
-progress <boolean> Report progress
-help <string list> Display long help (empty list), or help for given parameters keys
Use -help param1 [... paramN] to see detailed documentation of those parameters.
Examples:
otbcli_TensorflowModelTrain -source1.il spot6pms.tif -source1.placeholder x1 -source1.patchsizex 16 -source1.patchsizey 16 -source2.il labels.tif -source2.placeholder y1 -source2.patchsizex 1 -source2.patchsizex 1 -model.dir /tmp/my_saved_model/ -training.userplaceholders is_training=true dropout=0.2 -training.targetnodes optimizer -model.saveto /tmp/my_saved_model/variables/variables
As you can note, there is $OTB_TF_NSOURCES + 1 sources because we often need at least one more source for the reference data (e.g. terrain truth for land cover mapping).
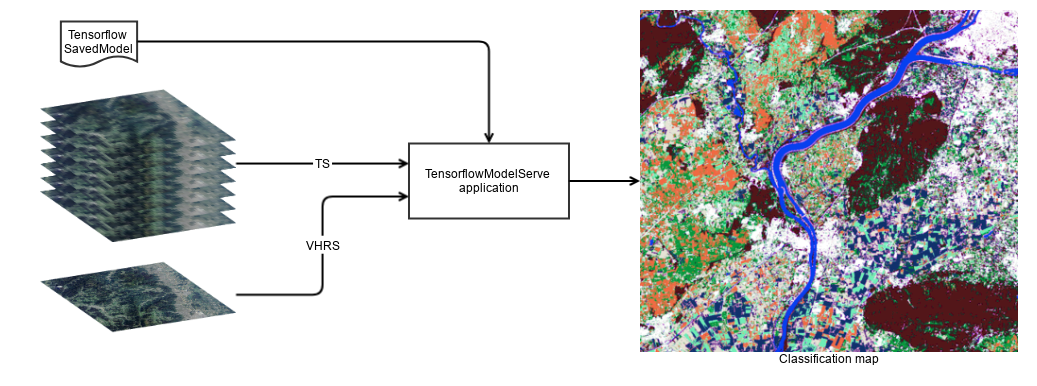
The TensorflowModelServe application perform model serving, it can be used to produce output raster with the desired tensors.
Thanks to the streaming mechanism, very large images can be produced.
The application uses the TensorflowModelFilter and a StreamingFilter to force the streaming of output.
This last can be optionally disabled by the user, if he prefers using the extended filenames to deal with chunk sizes.
However, it's still very useful when the application is used in other composites applications, or just without extended filename magic.
Some models can consume a lot of memory.
In addition, the native tiling strategy of OTB consists in strips but this might not always the best.
For Convolutional Neural Networks for instance, square tiles are more interesting because the padding required to perform the computation of one single strip of pixels induces to input a lot more pixels that to process the computation of one single tile of pixels.
So, this application takes in input one or multiple input sources (the number of input sources can be changed by setting the OTB_TF_NSOURCES to the desired number) and produce one output of the specified tensors.
The user is responsible of giving the receptive field and name of input placeholders, as well as the expression field, scale factor and name of output tensors.
The first input source (source1.il) corresponds to the reference input source.
As explained previously, the scale factor provided for the output tensors is related to this reference input source.
The user can ask for multiple output tensors, that will be stack along the channel dimension of the output raster.
However, if the sizes of those output tensors are not consistent (e.g. a different number of (x,y) elements), an exception will be thrown.
Multisource deep learning classifier using TensorFlow. Change the OTB_TF_NSOURCES environment variable to set the number of sources.
Parameters:
-source1 <group> Parameters for source #1
MISSING -source1.il <string list> Input image (or list to stack) for source #1 (mandatory)
MISSING -source1.rfieldx <int32> Input receptive field (width) for source #1 (mandatory)
MISSING -source1.rfieldy <int32> Input receptive field (height) for source #1 (mandatory)
MISSING -source1.placeholder <string> Name of the input placeholder for source #1 (mandatory)
-model <group> model parameters
MISSING -model.dir <string> TensorFlow model_save directory (mandatory)
-model.userplaceholders <string list> Additional single-valued placeholders. Supported types: int, float, bool. (optional, off by default)
-model.fullyconv <boolean> Fully convolutional (optional, off by default, default value is false)
-output <group> Output tensors parameters
-output.spcscale <float> The output spacing scale, related to the first input (mandatory, default value is 1)
MISSING -output.names <string list> Names of the output tensors (mandatory)
-output.efieldx <int32> The output expression field (width) (mandatory, default value is 1)
-output.efieldy <int32> The output expression field (height) (mandatory, default value is 1)
-optim <group> This group of parameters allows optimization of processing time
-optim.disabletiling <boolean> Disable tiling (optional, off by default, default value is false)
-optim.tilesizex <int32> Tile width used to stream the filter output (mandatory, default value is 16)
-optim.tilesizey <int32> Tile height used to stream the filter output (mandatory, default value is 16)
MISSING -out <string> [pixel] output image [pixel=uint8/uint16/int16/uint32/int32/float/double/cint16/cint32/cfloat/cdouble] (default value is float) (mandatory)
-progress <boolean> Report progress
-help <string list> Display long help (empty list), or help for given parameters keys
Use -help param1 [... paramN] to see detailed documentation of those parameters.
Examples:
otbcli_TensorflowModelServe -source1.il spot6pms.tif -source1.placeholder x1 -source1.rfieldx 16 -source1.rfieldy 16 -model.dir /tmp/my_saved_model/ -model.userplaceholders is_training=false dropout=0.0 -output.names out_predict1 out_proba1 -out "classif128tgt.tif?&streaming:type=tiled&streaming:sizemode=height&streaming:sizevalue=256"
Who has never dreamed to use classic classifiers performing on deep learning features? This is possible thank to two new applications that uses the existing training/classification applications of OTB:
TrainClassifierFromDeepFeatures: is a composite application that wire the TensorflowModelServe application output into the existing official TrainImagesClassifier application.
Train a classifier from deep net based features of an image and training vector data.
Parameters:
-source1 <group> Parameters for source 1
MISSING -source1.il <string list> Input image (or list to stack) for source #1 (mandatory)
MISSING -source1.rfieldx <int32> Input receptive field (width) for source #1 (mandatory)
MISSING -source1.rfieldy <int32> Input receptive field (height) for source #1 (mandatory)
MISSING -source1.placeholder <string> Name of the input placeholder for source #1 (mandatory)
-model <group> Deep net inputs parameters
MISSING -model.dir <string> TensorFlow model_save directory (mandatory)
-model.userplaceholders <string list> Additional single-valued placeholders. Supported types: int, float, bool. (optional, off by default)
-model.fullyconv <boolean> Fully convolutional (optional, off by default, default value is false)
-output <group> Deep net outputs parameters
-output.spcscale <float> The output spacing scale, related to the first input (mandatory, default value is 1)
MISSING -output.names <string list> Names of the output tensors (mandatory)
-output.efieldx <int32> The output expression field (width) (mandatory, default value is 1)
-output.efieldy <int32> The output expression field (height) (mandatory, default value is 1)
-optim <group> Processing time optimization
-optim.disabletiling <boolean> Disable tiling (optional, off by default, default value is false)
-optim.tilesizex <int32> Tile width used to stream the filter output (mandatory, default value is 16)
-optim.tilesizey <int32> Tile height used to stream the filter output (mandatory, default value is 16)
-ram <int32> Available RAM (Mb) (optional, off by default, default value is 128)
MISSING -vd <string list> Vector data for training (mandatory)
-valid <string list> Vector data for validation (optional, off by default)
MISSING -out <string> Output classification model (mandatory)
-confmatout <string> Output confusion matrix (optional, off by default)
-sample <group> Sampling parameters
-sample.mt <int32> Maximum training sample size per class (mandatory, default value is 1000)
-sample.mv <int32> Maximum validation sample size per class (mandatory, default value is 1000)
-sample.bm <int32> Bound sample number by minimum (mandatory, default value is 1)
-sample.vtr <float> Training and validation sample ratio (mandatory, default value is 0.5)
-sample.vfn <string> Field containing the class integer label for supervision (mandatory, no default value)
-elev <group> Elevation parameters
-elev.dem <string> DEM directory (optional, off by default)
-elev.geoid <string> Geoid File (optional, off by default)
-elev.default <float> Default elevation (mandatory, default value is 0)
-classifier <string> Classifier parameters [libsvm/boost/dt/gbt/ann/bayes/rf/knn/sharkrf/sharkkm] (mandatory, default value is libsvm)
-classifier.libsvm.k <string> SVM Kernel Type [linear/rbf/poly/sigmoid] (mandatory, default value is linear)
-classifier.libsvm.m <string> SVM Model Type [csvc/nusvc/oneclass] (mandatory, default value is csvc)
-classifier.libsvm.c <float> Cost parameter C (mandatory, default value is 1)
-classifier.libsvm.nu <float> Cost parameter Nu (mandatory, default value is 0.5)
-classifier.libsvm.opt <boolean> Parameters optimization (mandatory, default value is false)
-classifier.libsvm.prob <boolean> Probability estimation (mandatory, default value is false)
-classifier.boost.t <string> Boost Type [discrete/real/logit/gentle] (mandatory, default value is real)
-classifier.boost.w <int32> Weak count (mandatory, default value is 100)
-classifier.boost.r <float> Weight Trim Rate (mandatory, default value is 0.95)
-classifier.boost.m <int32> Maximum depth of the tree (mandatory, default value is 1)
-classifier.dt.max <int32> Maximum depth of the tree (mandatory, default value is 65535)
-classifier.dt.min <int32> Minimum number of samples in each node (mandatory, default value is 10)
-classifier.dt.ra <float> Termination criteria for regression tree (mandatory, default value is 0.01)
-classifier.dt.cat <int32> Cluster possible values of a categorical variable into K <= cat clusters to find a suboptimal split (mandatory, default value is 10)
-classifier.dt.f <int32> K-fold cross-validations (mandatory, default value is 10)
-classifier.dt.r <boolean> Set Use1seRule flag to false (mandatory, default value is false)
-classifier.dt.t <boolean> Set TruncatePrunedTree flag to false (mandatory, default value is false)
-classifier.gbt.w <int32> Number of boosting algorithm iterations (mandatory, default value is 200)
-classifier.gbt.s <float> Regularization parameter (mandatory, default value is 0.01)
-classifier.gbt.p <float> Portion of the whole training set used for each algorithm iteration (mandatory, default value is 0.8)
-classifier.gbt.max <int32> Maximum depth of the tree (mandatory, default value is 3)
-classifier.ann.t <string> Train Method Type [back/reg] (mandatory, default value is reg)
-classifier.ann.sizes <string list> Number of neurons in each intermediate layer (mandatory)
-classifier.ann.f <string> Neuron activation function type [ident/sig/gau] (mandatory, default value is sig)
-classifier.ann.a <float> Alpha parameter of the activation function (mandatory, default value is 1)
-classifier.ann.b <float> Beta parameter of the activation function (mandatory, default value is 1)
-classifier.ann.bpdw <float> Strength of the weight gradient term in the BACKPROP method (mandatory, default value is 0.1)
-classifier.ann.bpms <float> Strength of the momentum term (the difference between weights on the 2 previous iterations) (mandatory, default value is 0.1)
-classifier.ann.rdw <float> Initial value Delta_0 of update-values Delta_{ij} in RPROP method (mandatory, default value is 0.1)
-classifier.ann.rdwm <float> Update-values lower limit Delta_{min} in RPROP method (mandatory, default value is 1e-07)
-classifier.ann.term <string> Termination criteria [iter/eps/all] (mandatory, default value is all)
-classifier.ann.eps <float> Epsilon value used in the Termination criteria (mandatory, default value is 0.01)
-classifier.ann.iter <int32> Maximum number of iterations used in the Termination criteria (mandatory, default value is 1000)
-classifier.rf.max <int32> Maximum depth of the tree (mandatory, default value is 5)
-classifier.rf.min <int32> Minimum number of samples in each node (mandatory, default value is 10)
-classifier.rf.ra <float> Termination Criteria for regression tree (mandatory, default value is 0)
-classifier.rf.cat <int32> Cluster possible values of a categorical variable into K <= cat clusters to find a suboptimal split (mandatory, default value is 10)
-classifier.rf.var <int32> Size of the randomly selected subset of features at each tree node (mandatory, default value is 0)
-classifier.rf.nbtrees <int32> Maximum number of trees in the forest (mandatory, default value is 100)
-classifier.rf.acc <float> Sufficient accuracy (OOB error) (mandatory, default value is 0.01)
-classifier.knn.k <int32> Number of Neighbors (mandatory, default value is 32)
-classifier.sharkrf.nbtrees <int32> Maximum number of trees in the forest (mandatory, default value is 100)
-classifier.sharkrf.nodesize <int32> Min size of the node for a split (mandatory, default value is 25)
-classifier.sharkrf.mtry <int32> Number of features tested at each node (mandatory, default value is 0)
-classifier.sharkrf.oobr <float> Out of bound ratio (mandatory, default value is 0.66)
-classifier.sharkkm.maxiter <int32> Maximum number of iteration for the kmeans algorithm. (mandatory, default value is 10)
-classifier.sharkkm.k <int32> The number of class used for the kmeans algorithm. (mandatory, default value is 2)
-rand <int32> User defined random seed (optional, off by default)
-inxml <string> Load otb application from xml file (optional, off by default)
-progress <boolean> Report progress
-help <string list> Display long help (empty list), or help for given parameters keys
Use -help param1 [... paramN] to see detailed documentation of those parameters.
Examples:
None
ImageClassifierFromDeepFeatures same approach with the official ImageClassifier.
Classify image using features from a deep net and an OTB machine learning classification model
Parameters:
-source1 <group> Parameters for source 1
MISSING -source1.il <string list> Input image (or list to stack) for source #1 (mandatory)
MISSING -source1.rfieldx <int32> Input receptive field (width) for source #1 (mandatory)
MISSING -source1.rfieldy <int32> Input receptive field (height) for source #1 (mandatory)
MISSING -source1.placeholder <string> Name of the input placeholder for source #1 (mandatory)
-deepmodel <group> Deep net model parameters
MISSING -deepmodel.dir <string> TensorFlow model_save directory (mandatory)
-deepmodel.userplaceholders <string list> Additional single-valued placeholders. Supported types: int, float, bool. (optional, off by default)
-deepmodel.fullyconv <boolean> Fully convolutional (optional, off by default, default value is false)
-output <group> Deep net outputs parameters
-output.spcscale <float> The output spacing scale, related to the first input (mandatory, default value is 1)
MISSING -output.names <string list> Names of the output tensors (mandatory)
-output.efieldx <int32> The output expression field (width) (mandatory, default value is 1)
-output.efieldy <int32> The output expression field (height) (mandatory, default value is 1)
-optim <group> This group of parameters allows optimization of processing time
-optim.disabletiling <boolean> Disable tiling (optional, off by default, default value is false)
-optim.tilesizex <int32> Tile width used to stream the filter output (mandatory, default value is 16)
-optim.tilesizey <int32> Tile height used to stream the filter output (mandatory, default value is 16)
MISSING -model <string> Model file (mandatory)
-imstat <string> Statistics file (optional, off by default)
-nodatalabel <int32> Label mask value (optional, off by default, default value is 0)
MISSING -out <string> [pixel] Output image [pixel=uint8/uint16/int16/uint32/int32/float/double/cint16/cint32/cfloat/cdouble] (default value is uint8) (mandatory)
-confmap <string> [pixel] Confidence map image [pixel=uint8/uint16/int16/uint32/int32/float/double/cint16/cint32/cfloat/cdouble] (default value is double) (optional, off by default)
-ram <int32> Ram (optional, off by default, default value is 128)
-inxml <string> Load otb application from xml file (optional, off by default)
-progress <boolean> Report progress
-help <string list> Display long help (empty list), or help for given parameters keys
Use -help param1 [... paramN] to see detailed documentation of those parameters.
Examples:
None
Note that you can still set the OTB_TF_NSOURCES environment variable.
Here we will try to provide a simple example of doing a classification using a deep net that performs on one single VHR image. Our data set consists in one Spot-7 image, spot7.tif, and a training vector data, terrain_truth.shp that describes sparsely forest / non-forest polygons. First, we compute statistics of the vector data : how many points can we sample inside objects, and how many objects in each class. We use the PolygonClassStatistics application of OTB.
otbcli_PolygonClassStatistics -vec terrain_truth.shp -field class -in spot7.tif -out vec_stats.xml
Then, we will select some samples with the SampleSelection application of the existing machine learning framework of OTB. Since the terrain truth is sparse, we want to sample randomly points in polygons with the default strategy of the SampleSelection OTB application.
otbcli_SampleSelection -in spot7.tif -vec terrain_truth.shp -instats vec_stats.xml -field class -out points.shp
Now we extract the patches with the PatchesExtraction application. We want to produce one image of 16x16 patches, and one image for the corresponding labels.
otbcli_PatchesExtraction -source1.il spot7.tif -source1.patchsizex 16 -source1.patchsizey 16 -vec points.shp -field class -source1.out samp_labels.tif -outpatches samp_patches.tif
Now we have two images for patches and labels.
We can split them to distinguish test/validation groups (with the ExtractROI application for instance).
But here, we will just perform some fine tuning of our model.
The SavedModel is located in the outmodel directory.
Our model is quite basic: it has two input placeholders, x1 and y1 respectively for input patches (with size 16x16) and input reference labels (with size 1x1).
We named prediction the tensor that predict the labels and the optimizer that perform the stochastic gradient descent is an operator named optimizer.
We perform the fine tuning and we export the new model variables directly in the outmodel/variables folder, overwritting the existing variables of the model.
We use the TensorflowModelTrain application to perform the training of this existing model.
otbcli_TensorflowModelTrain -model.dir /path/to/oursavedmodel -training.targetnodesnames optimizer -training.source1.il samp_patches.tif -training.source1.patchsizex 16 -training.source1.patchsizey 16 -training.source1.placeholder x1 -training.source2.il samp_labels.tif -training.source2.patchsizex 1 -training.source2.patchsizey 1 -training.source2.placeholder y1 -model.saveto /path/to/oursavedmodel/variables/variables
Note that we could also have performed validation in this step. In this case, the validation.source2.placeholder would be different than the training.source2.placeholder, and would be prediction. This way, the program know what is the target tensor to evaluate.
After this step, we use the trained model to produce the entire map of forest over the whole Spot-7 image. For this, we use the TensorflowModelServe application to produce the prediction tensor output for the entire image.
otbcli_TensorflowModelServe -source1.il spot7.tif -source1.placeholder x1 -source1.rfieldx 16 -source1.rfieldy 16 -model.dir /path/to/oursavedmodel -output.names prediction -out map.tif uint8
In the python folder are provided some ready-to-use deep networks, with documentation and scientific references.
Feel free to contribute with your own architecture!
A tutorial is available at MDL4EO's blog
A full tutorial is available in this book. About 160 pages explain how to use OTBTF (with also OTB, QGIS and TensorFlow) for landcover mapping (patch-based image classification, fully convolutional models, hybrid deep networks X random forest classifiers, semantic segmentation from OSM data, image restoration with joint SAR/Optical sensors).