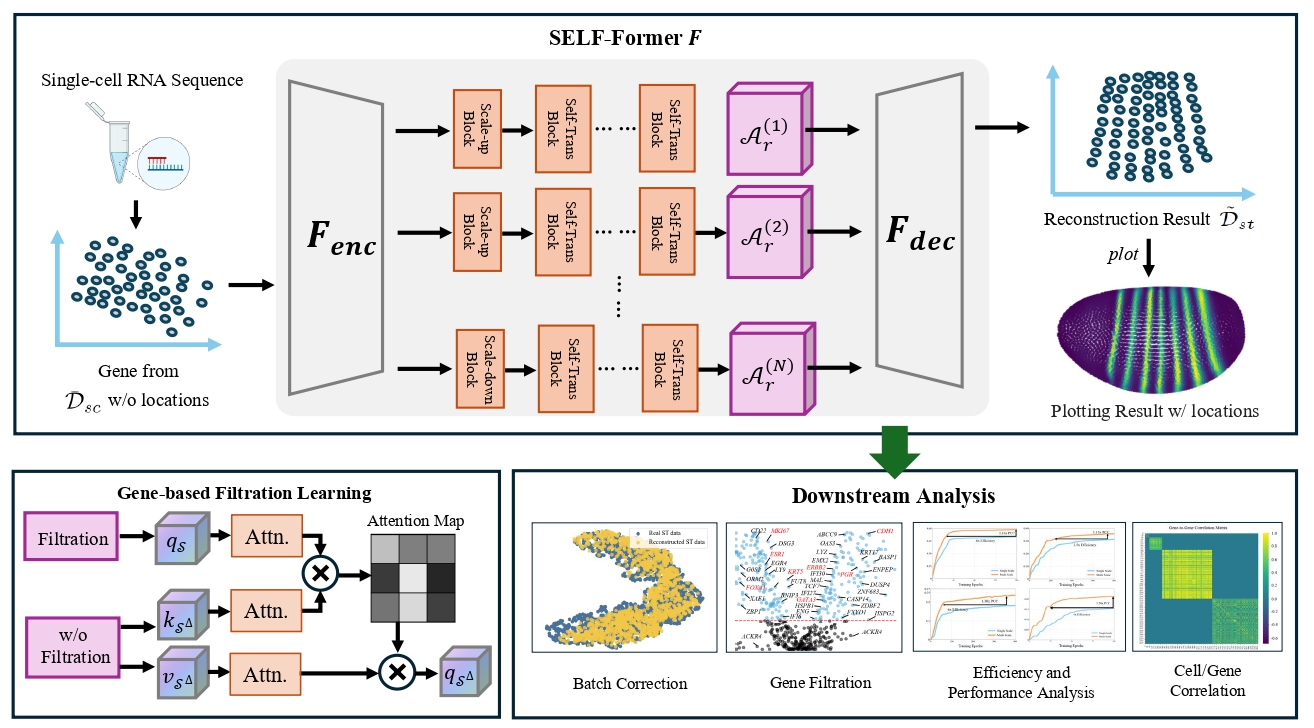
We developed SELF-Former, a transformer-based framework that utilizes multi-scale structures to learn gene representations while designing spatial correlation constraints for the reconstruction of corresponding spatial transcriptomics (ST) data.
SELF-Former is a novel framework that leverages the power of transformers to capture multi-scale gene representations. By incorporating spatial correlation constraints, SELF-Former effectively reconstructs spatial transcriptomics data from single-cell RNA sequencing (scRNA-seq) data. This approach enhances the accuracy and resolution of spatial gene expression patterns, providing a valuable tool for spatial omics research.
- In STARmap dataset, set hidden dimension to 2048, 1024, 1024, epoch to 400.
- In BRCA dataset, set hidden dimension to 8192, 4096, 4096, epoch to 100.
We provide training model code for STARmap and BRCA, as well as a tutorial for the visualization code.
The specific resulet settings follow those in the We provide training code for the corresponding datasets, which can be used to validate the test results mentioned in Spatial Benchmark and obtain the corresponding Excel data. Additionally, we provide the corresponding test results at ckpt Dropbox (metrics) and ckpt Dropbox (reconstructed results).
Create and activate the conda environment, then install required packages:
conda create -n SELF-Former python=3.6
conda activate SELF-Former
pip install -r requirements.txt