sv-channels
sv-channels is a Deep Learning workflow for calling structural variants (SVs) in short read alignment data using one-dimensional Convolutional Neural Networks (CNN). It has been tested on a benchmark dataset with three cell lines: two samples (NA12878 and NA24385) from the Genome in a Bottle consortium and one synthetic diploid sample (CHM1_CHM13).
sv-channels supports five SV types:
- deletions (DEL)
- insertions (INS)
- inversions (INV)
- tandem duplications (DUP)
- inter-chromosomal translocations (CTX)
The workflow includes the following key steps:
Transform read alignments into channels
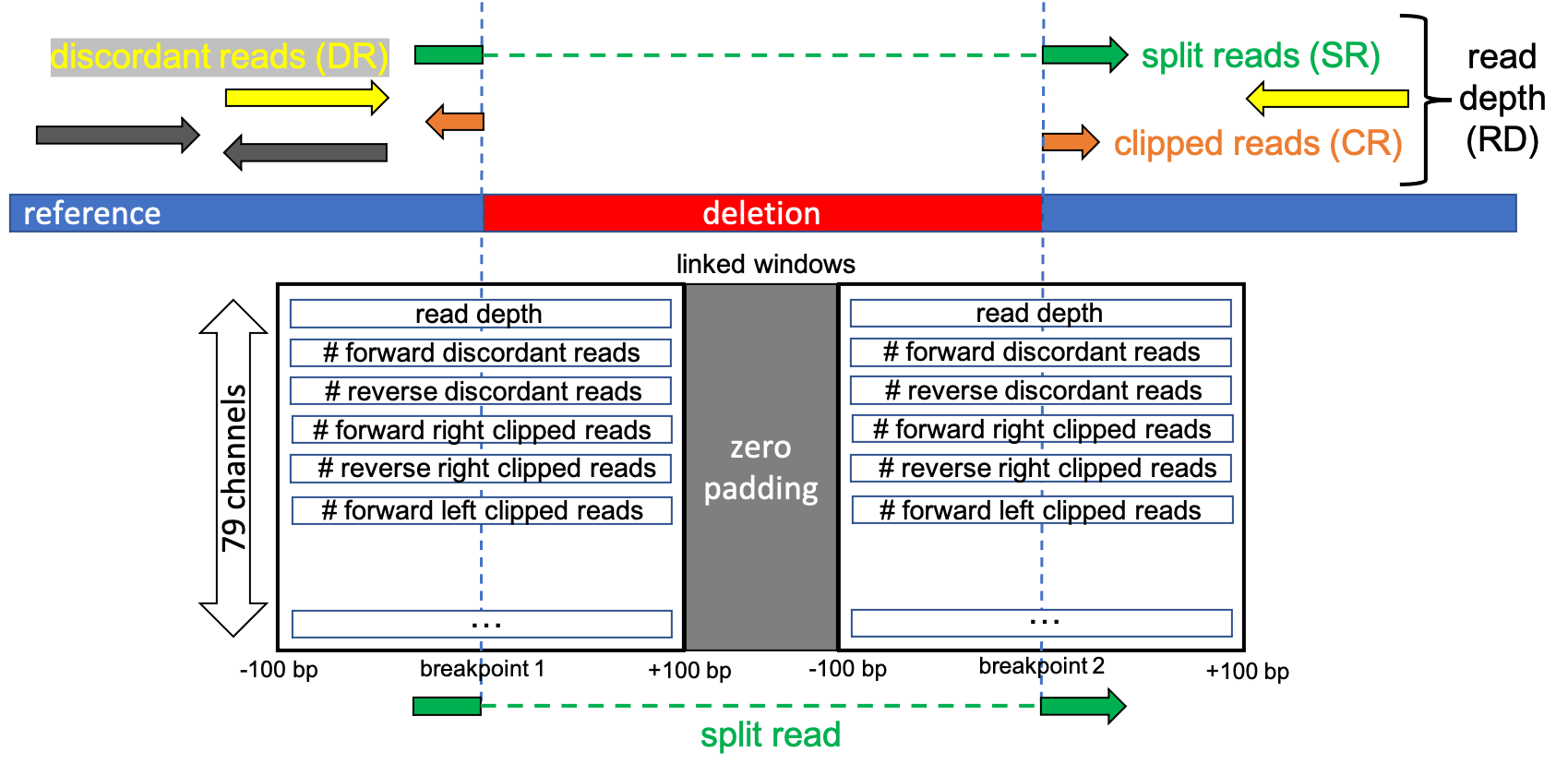
First, split read positions are extracted from the BAM files as candidate regions for SV breakpoints. For each pair of split read positions (rightmost position of the first split part and leftmost position of the second split part) a 2D Numpy array called window is constructed. The shape of a window is [window_size, number_of_channels], where the genomic interval encompassing the window is centered on the split read position with a context of [-100 bp, +100 bp) for a window_size of 200 bp. From all the reads overlapping this genomic interval and from the relative segment subsequence of the reference sequence 79 (number_of_channels) channels are constructed, where each channel encode a signal that can be used for SV calling. The list of channels can be found here. The two windows are joined as linked-windows with a zero padding 2D array of shape [10, number_of_channels] in between to avoid artifacts related to the CNN kernel in the part at the interface between the two windows. The linked-windows are labelled as SV when the split read positions overlap the SV callset used as the ground truth and noSV otherwise, where SV is either DEL,INS,INV,DUP or CTX according to the SV type.
Model training
The labelled linked-windows are used to train a 1D CNN to learn to classify them as either SV or noSV. Two cross-validation strategies are possible: 10-fold cross-validation and cross-validation by chromosome, where one chromosome is used as the test set and the other chromosomes as the training set.
SV calling with a trained model
Once a trained model is generated and the BAM file for the test set is converted into linked-windows, the SV calling is performed using the predict.py script.
Dependencies
- Conda
- listed in
environment.yaml
1. Clone this repo.
git clone https://github.com/GooglingTheCancerGenome/sv-channels.git
cd sv-channels2. Install dependencies.
# download Miniconda3 installer
wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh -O miniconda.sh
# install Conda (respond by 'yes')
bash miniconda.sh
# update Conda
conda update -y conda
# create & activate new env with installed deps
conda env create -n sv-channels -f environment.yaml
conda activate sv-channels3. Execution.
- input:
- output:
- SV callset in VCF format
SCH=local # gridengine or slurm
BAM=data/test.bam
SEQIDS="12,22"
SVTYPES="INV,DEL,INS,INV,DUP,CTX"
./run.sh $SCH $BAM $SEQIDS $SVTYPES # run jobs locally or on a compute clusterContributing
If you want to contribute to the development of sv-channels, have a look at the CONTRIBUTING.md.
License
Copyright (c) 2020, Netherlands eScience Center
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.