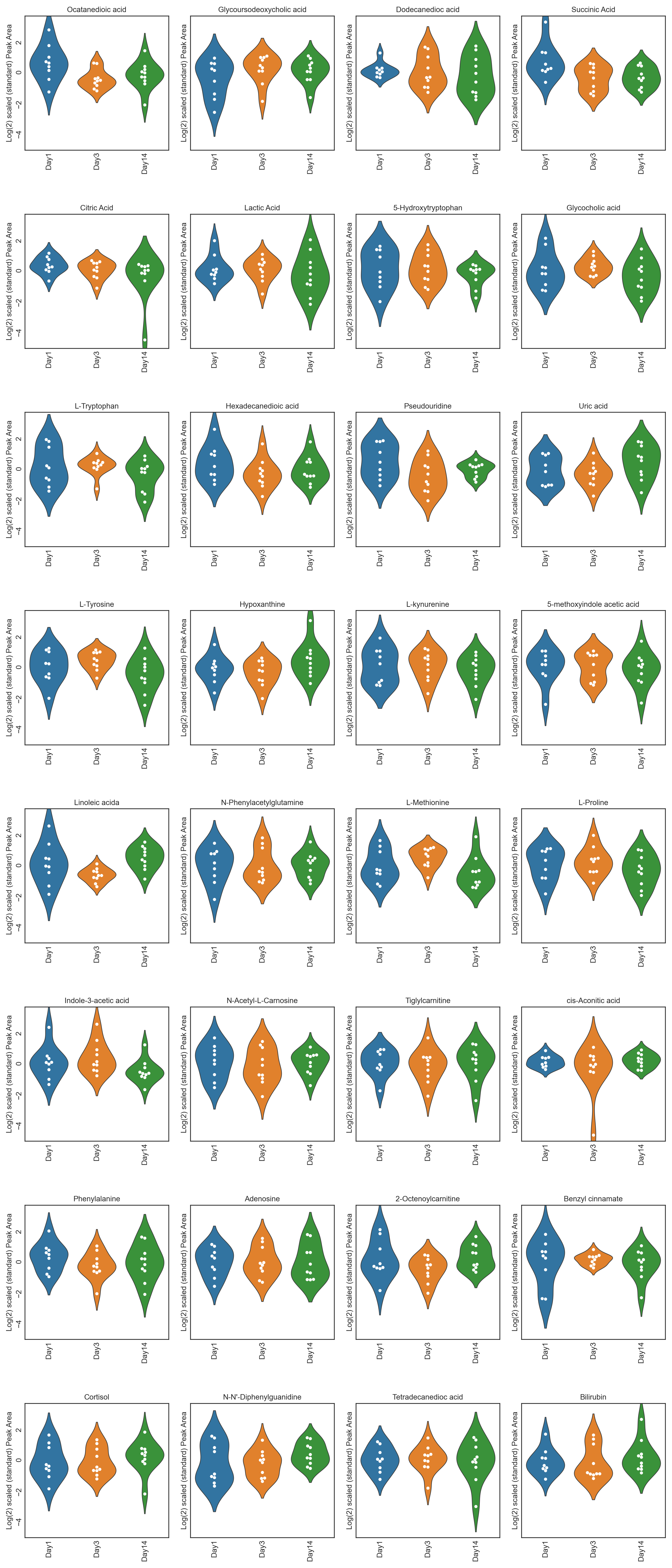
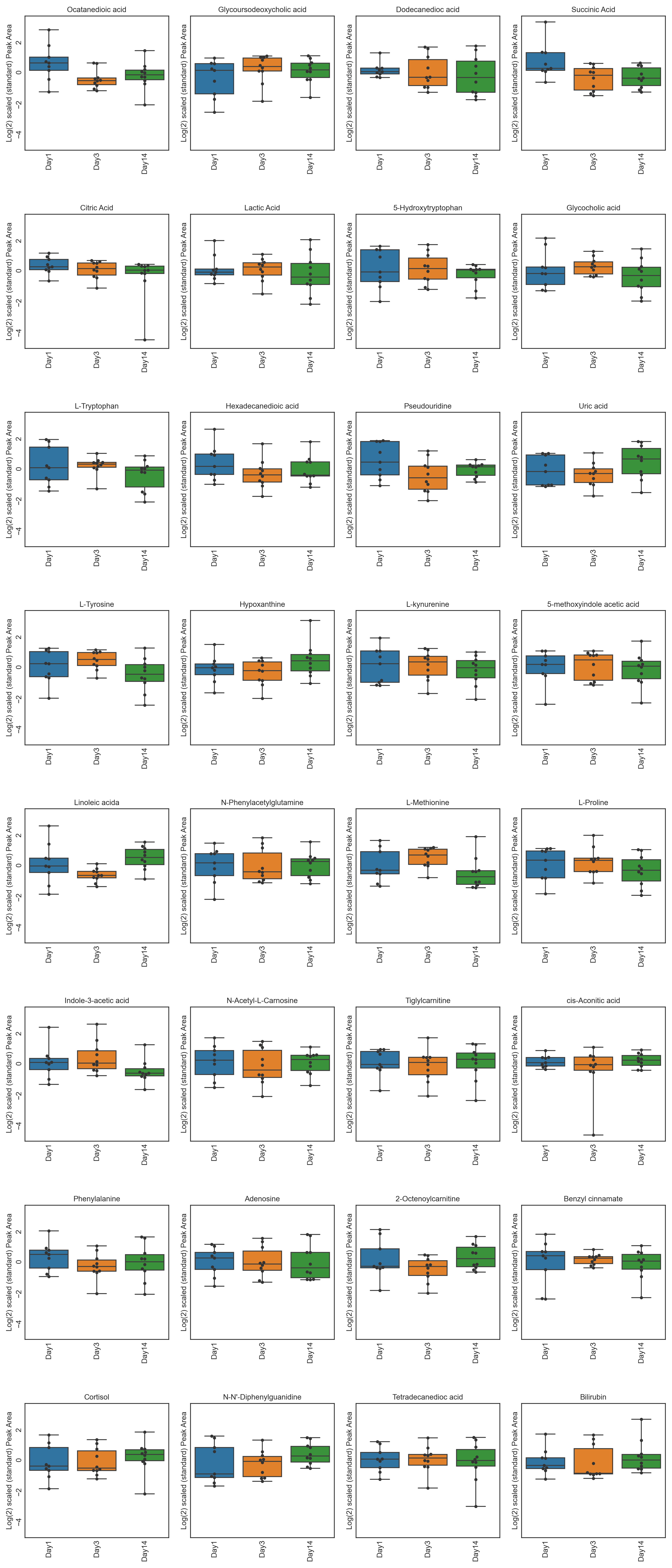
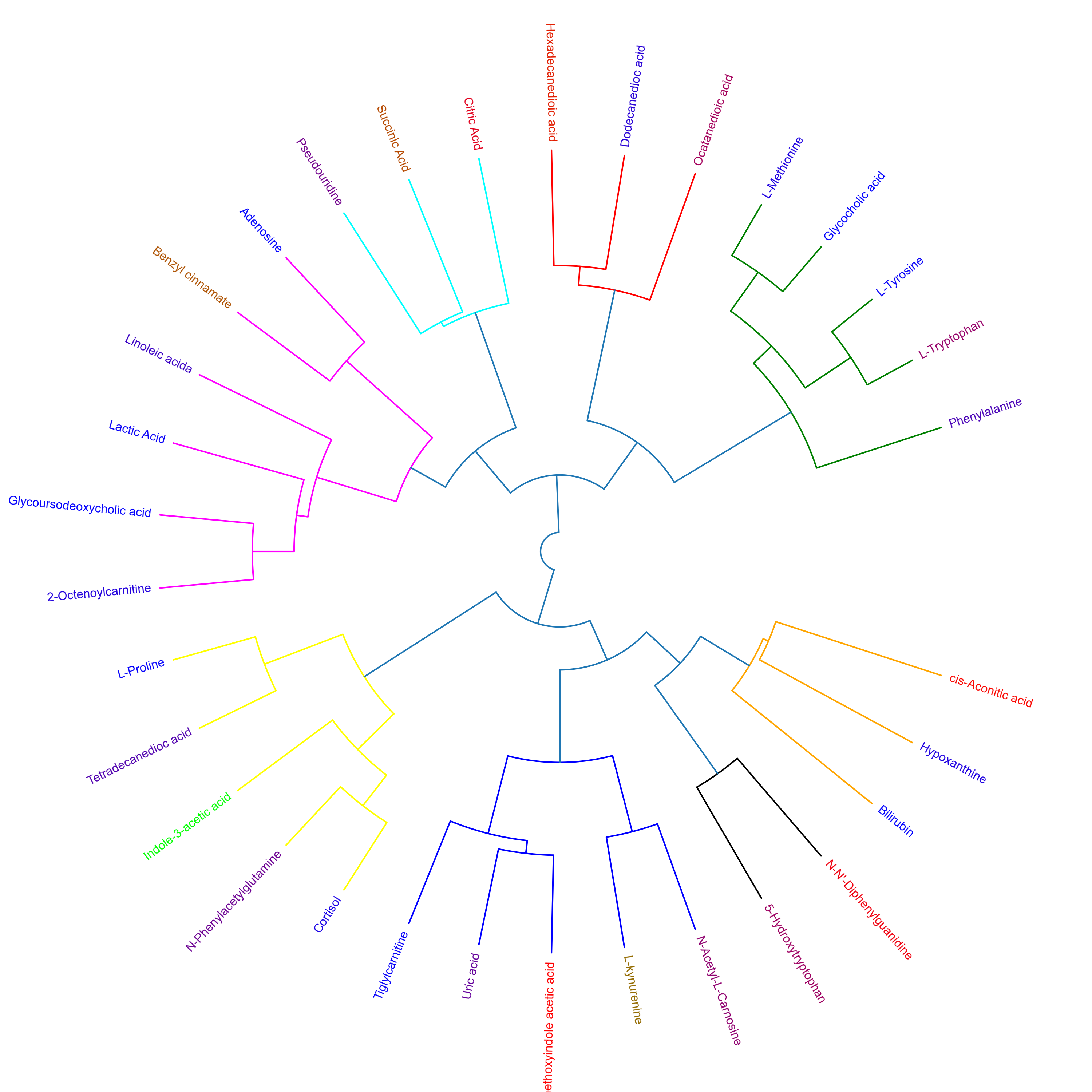
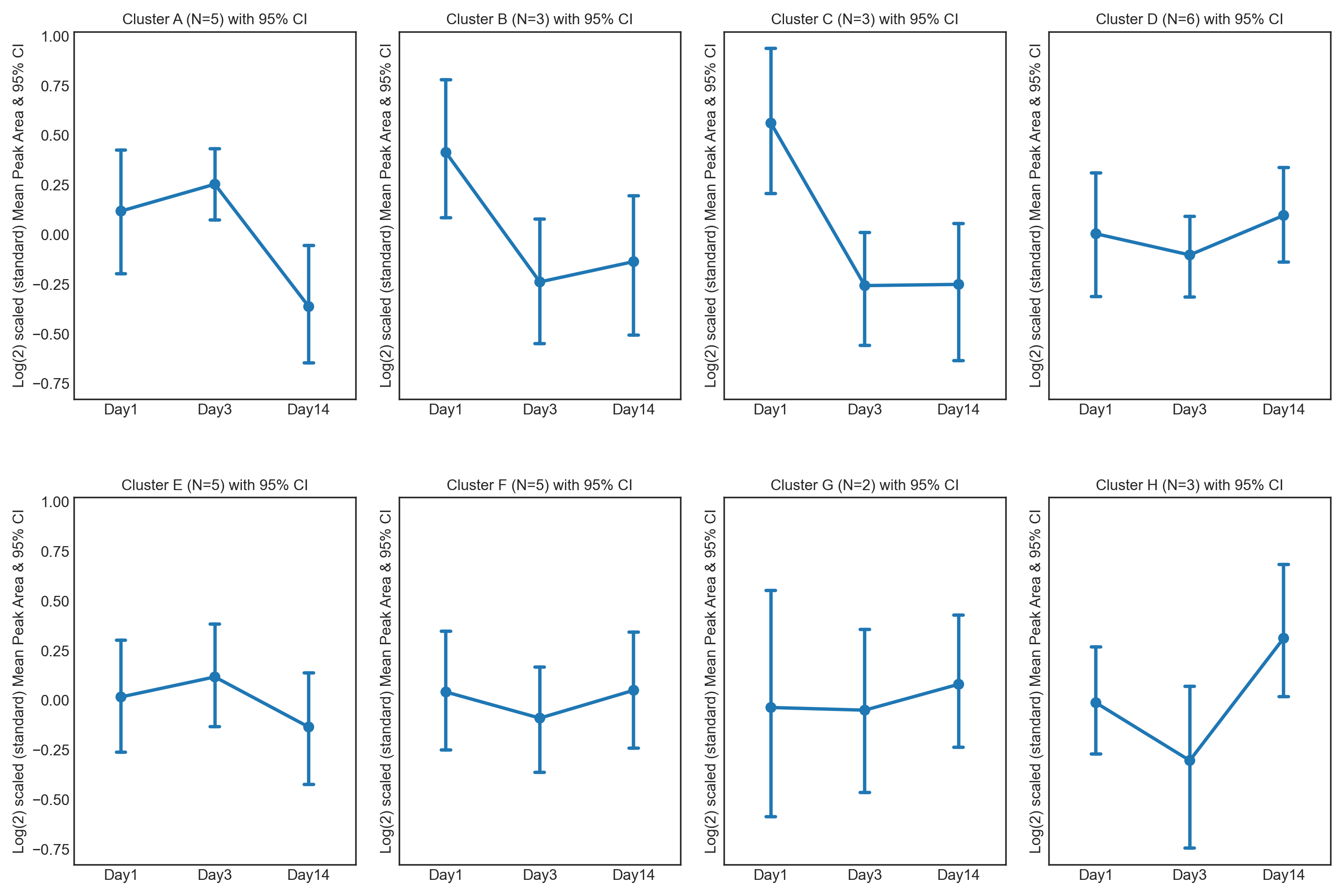
This repository contains twelve different tutorials spanning two case studies, explaining visualisations, from PCA, PCA loadings lollipop plots, PCoA, Hierarchical Clustered Heatmaps (HCH), Polar Dendrograms, Peak area plots, Spring-embedded network plots, and Hierarchical Edge Bundle plots, with included statistical analysis. Tutorials 1.1 to 1.6 cover a dataset from (Lawler et al. (2018)) published in Experimental Physiology, while tutorials 2.1 to 2.6 cover a dataset from (Chan, A, et al. (2016)) published in British Journal of Cancer. The tutorials stress the importance of multivariate analysis and provide multiple options for statistical analysis and multivariate visualisation. Each tutorial can be downloaded and run locally through Jupyter Notebook or run on the cloud through Binder.
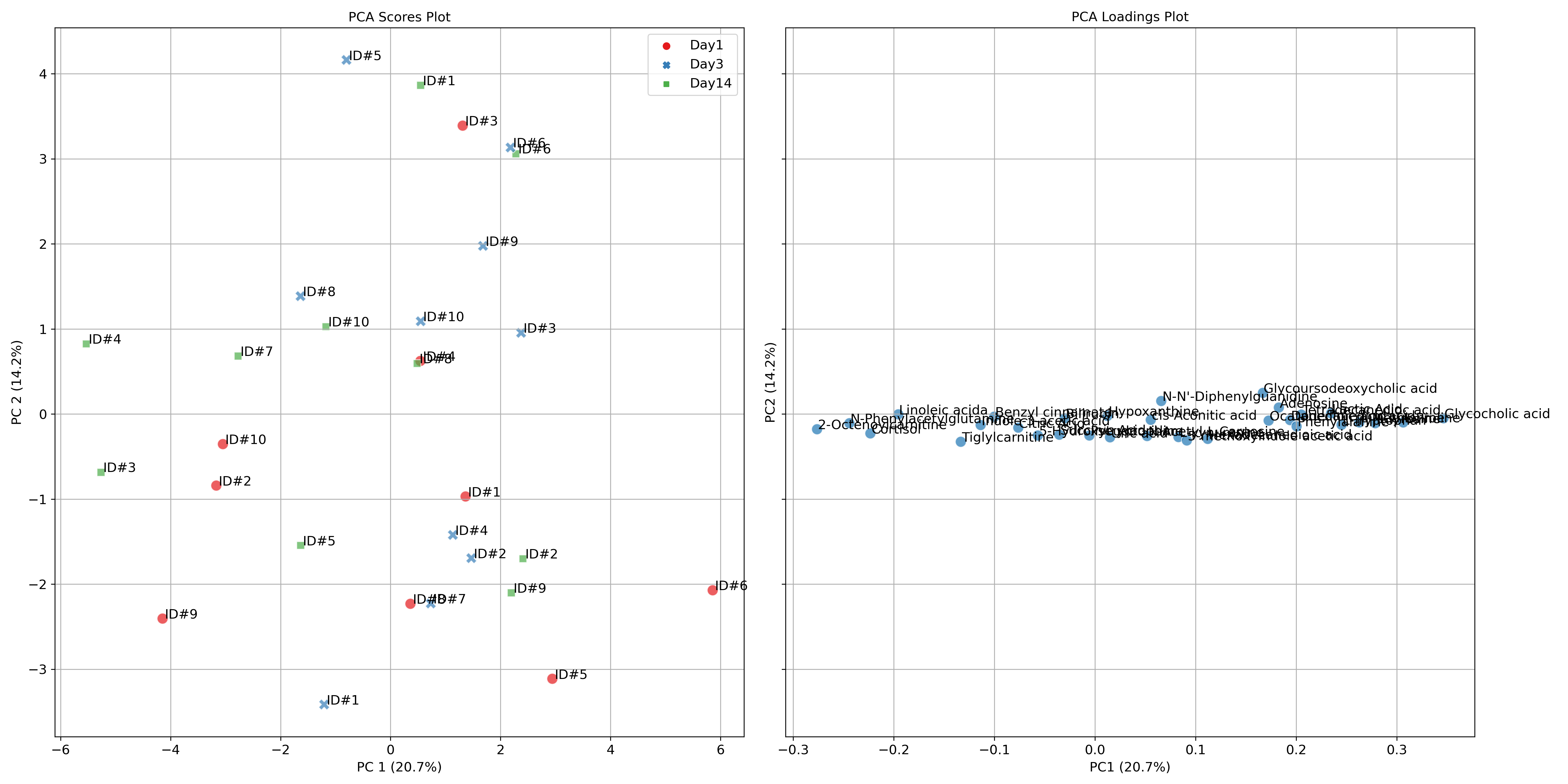
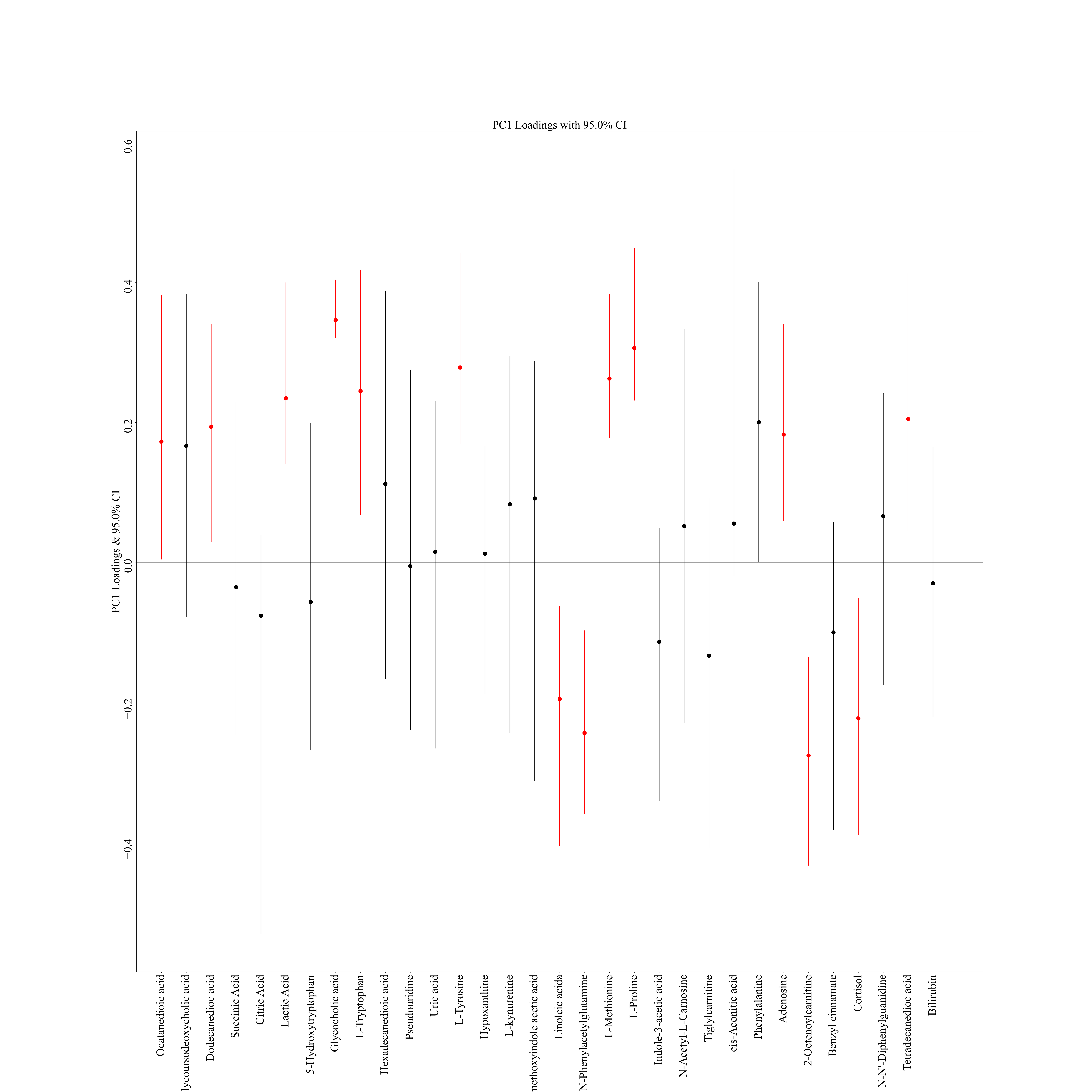
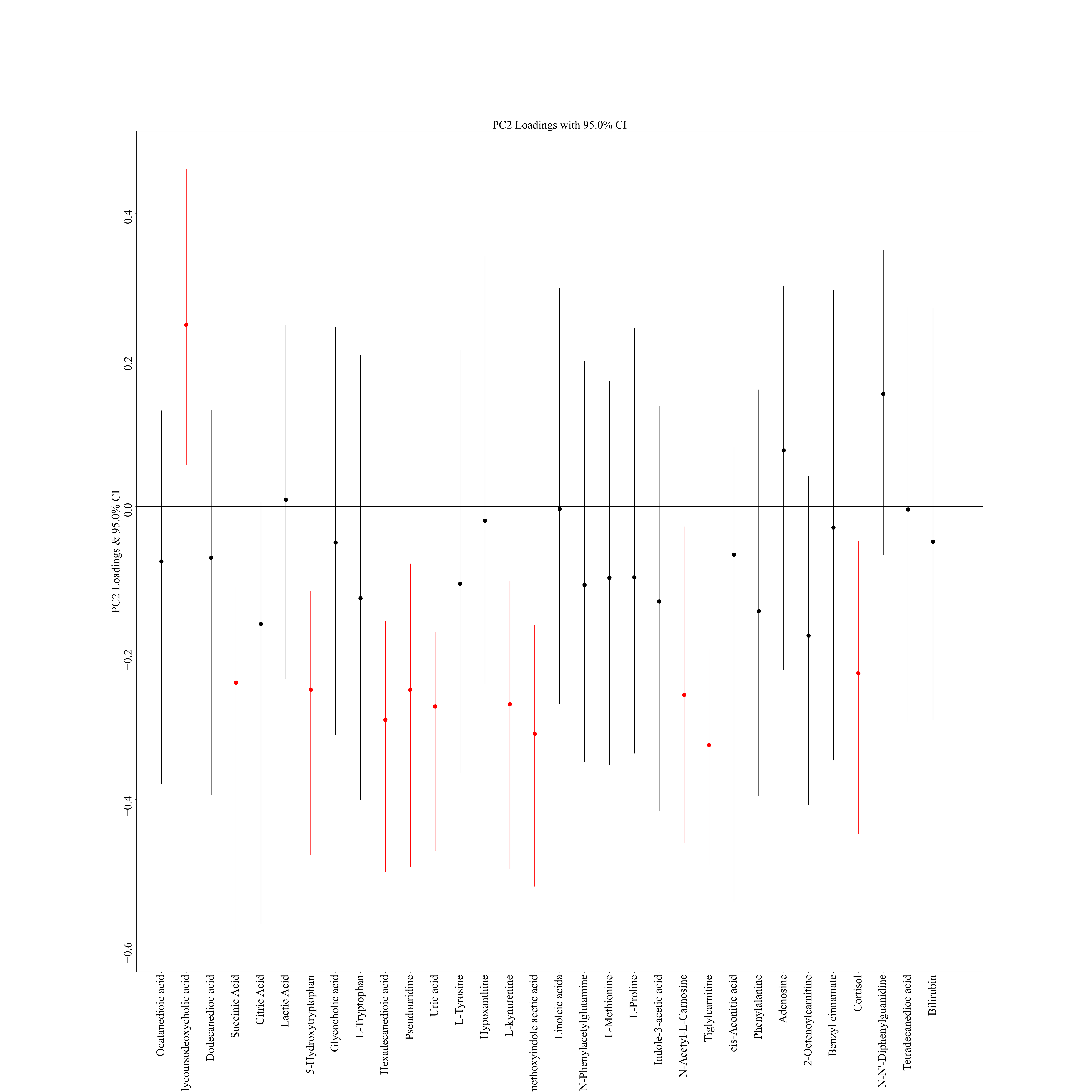
1.1. Athlete performance at high altitude - Principal Component Analysis (PCA) with loadings plot and PCA lollipop plots
1.2. Athlete performance at high altitude - Principal Coordinate Analysis (PCoA)
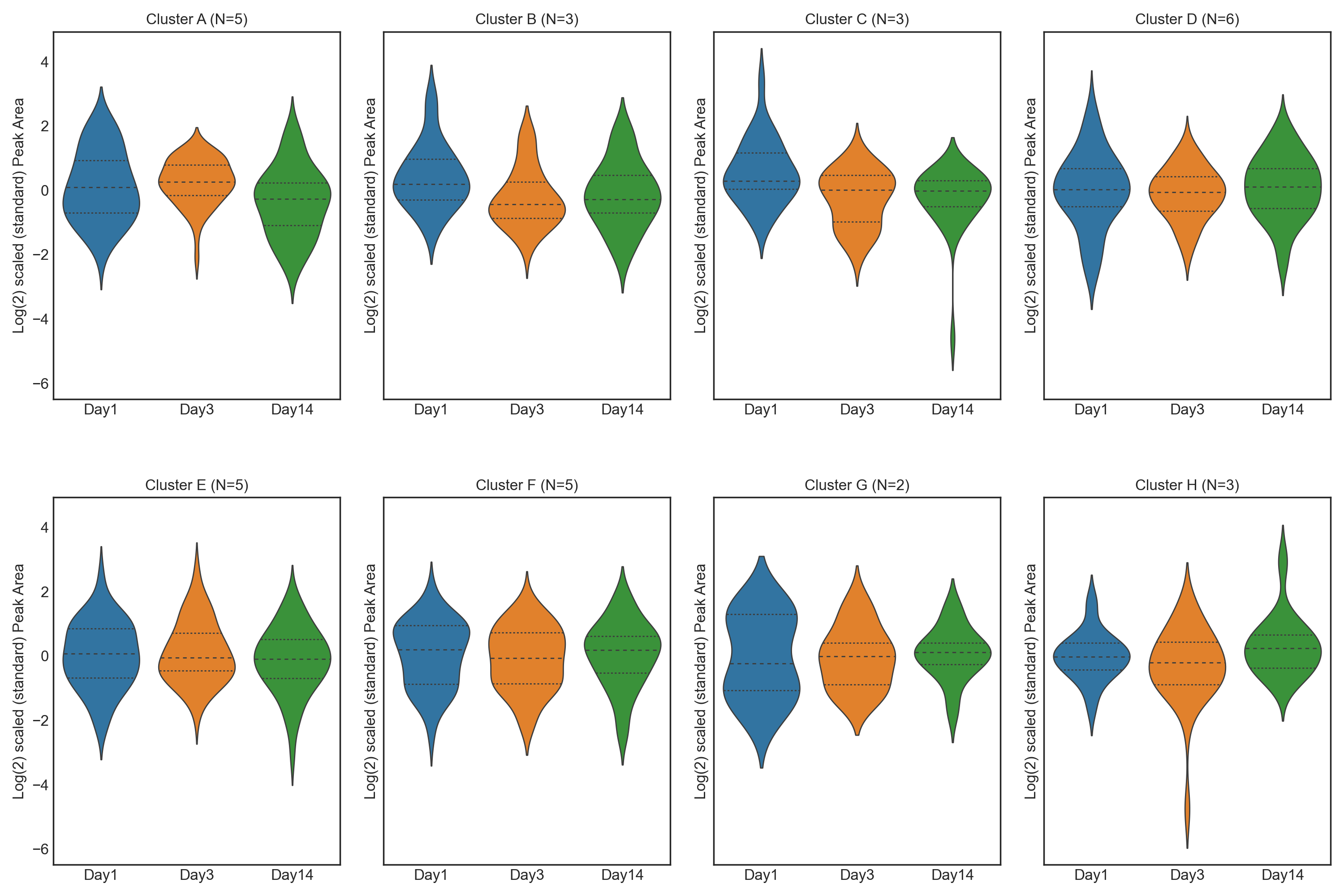
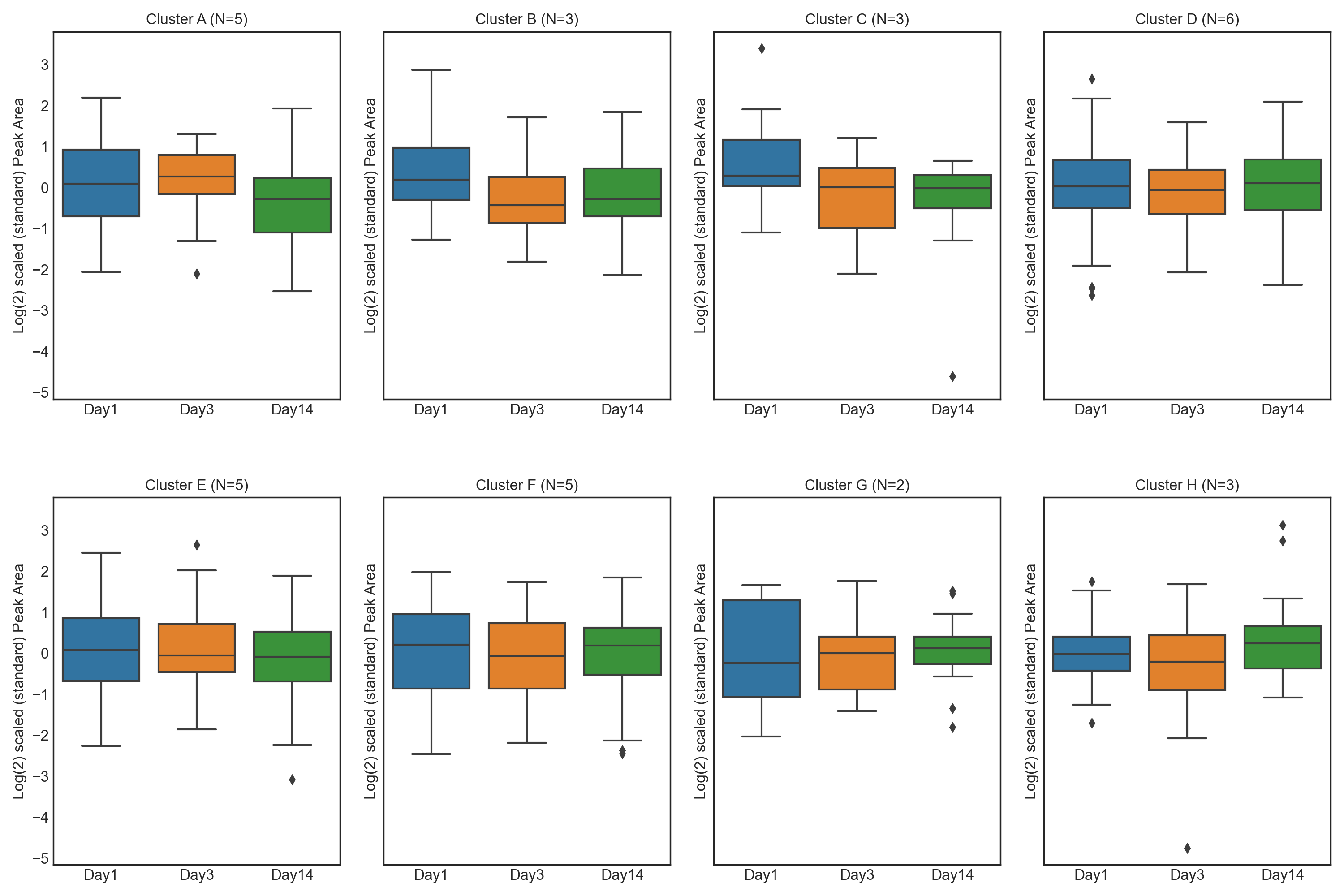
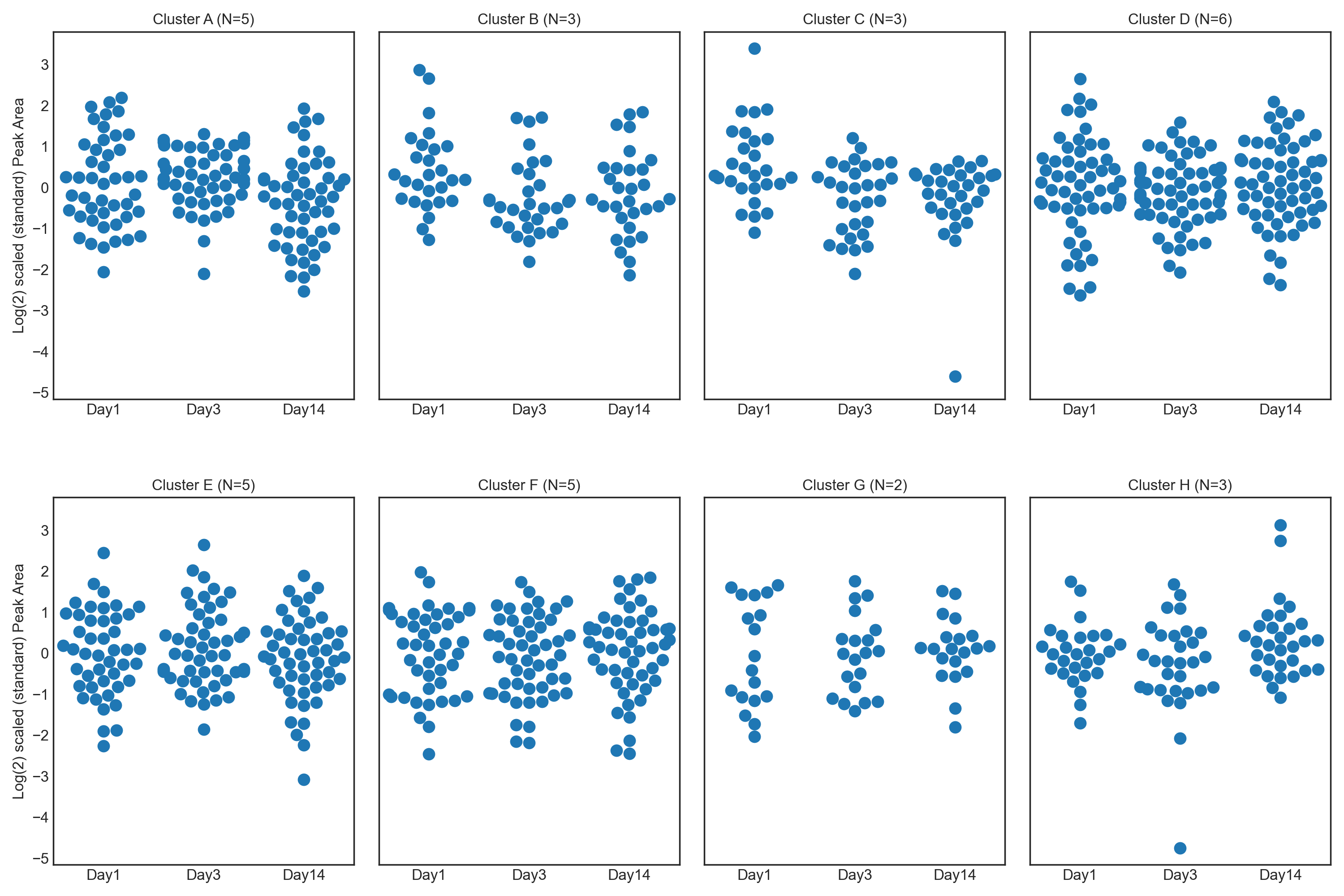
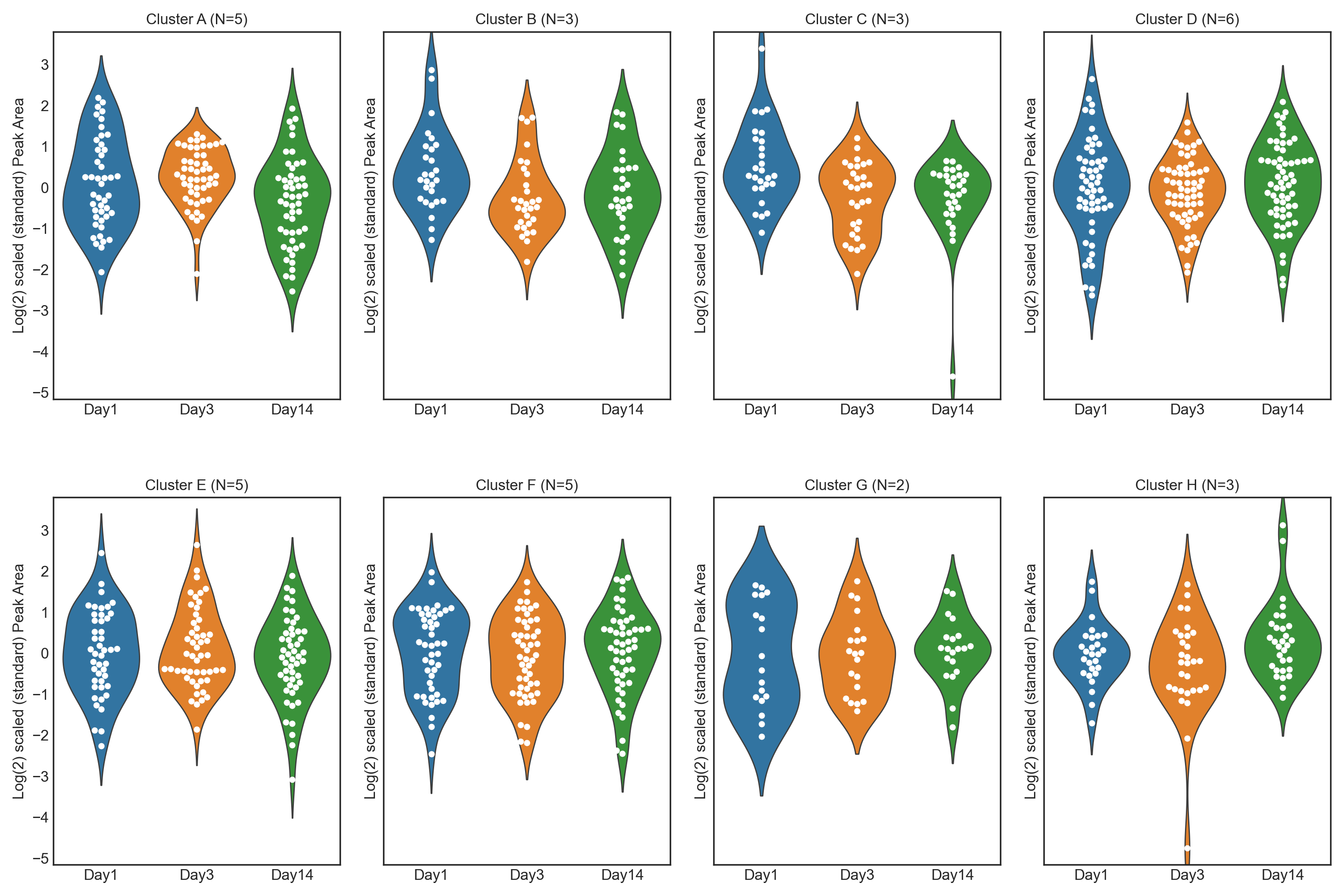
1.3. Athlete performance at high altitude - Feature Plot
1.4. Athlete performance at high altitude - Hierarchical Clustered Heatmap
1.5. Athlete performance at high altitude - Polar Dendrogram
1.6. Athlete performance at high altitude - Spring-embedded network
1.7. Athlete performance at high altitude - Hierarchical Edge Bundle
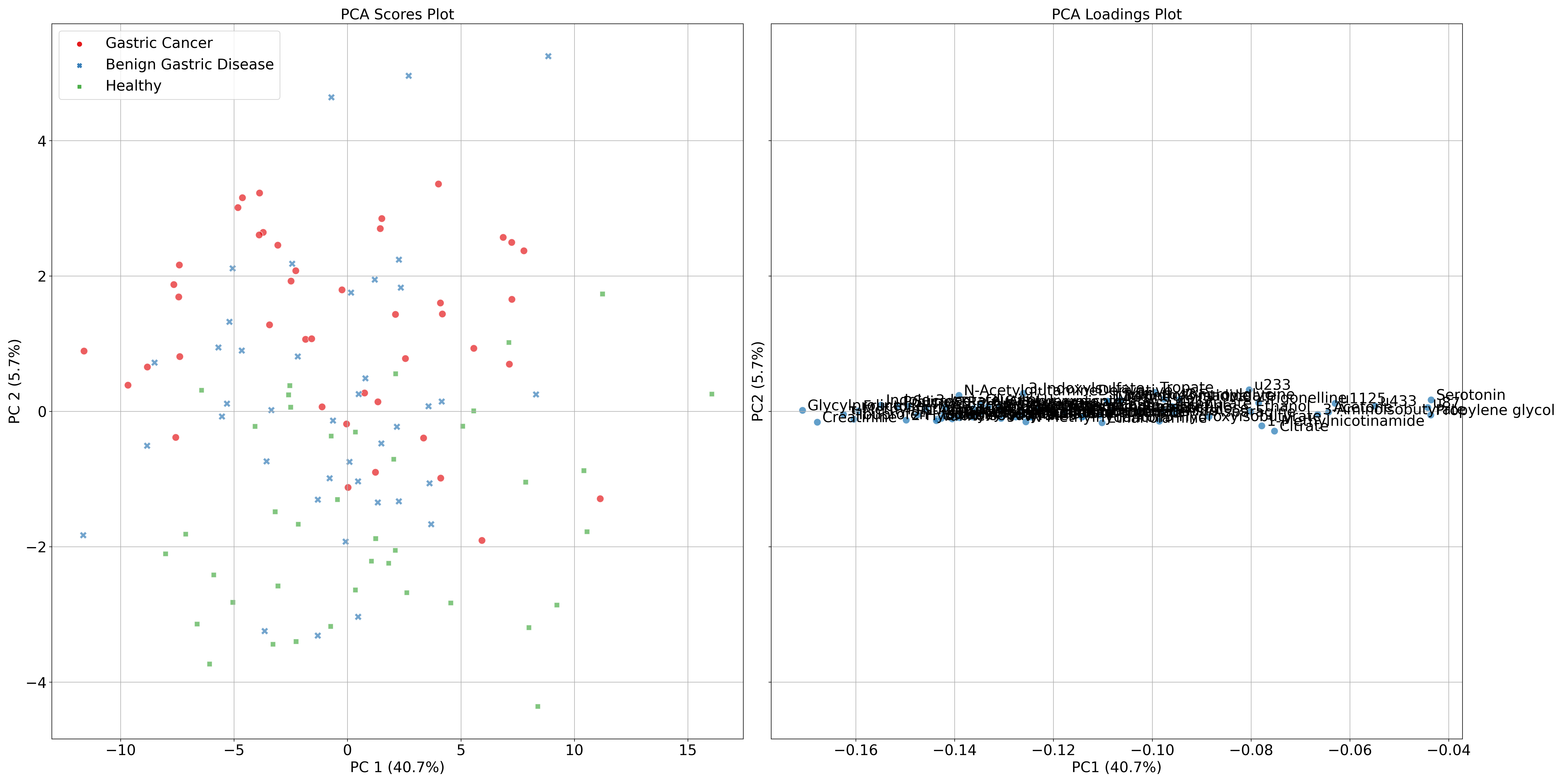
2.1. Gastric Cancer - Principal Component Analysis (PCA) with loadings plot and PCA lollipop plots
2.2. Gastric Cancer - Principal Coordinate Analysis (PCoA)
2.3. Gastric Cancer - Feature Plot
2.4. Gastric Cancer - Hierarchical Clustered Heatmap
2.5. Gastric Cancer - Polar Dendrogram
2.6. Gastric Cancer - Spring-embedded network
2.7. Gastric Cancer - Hierarchical Edge Bundle
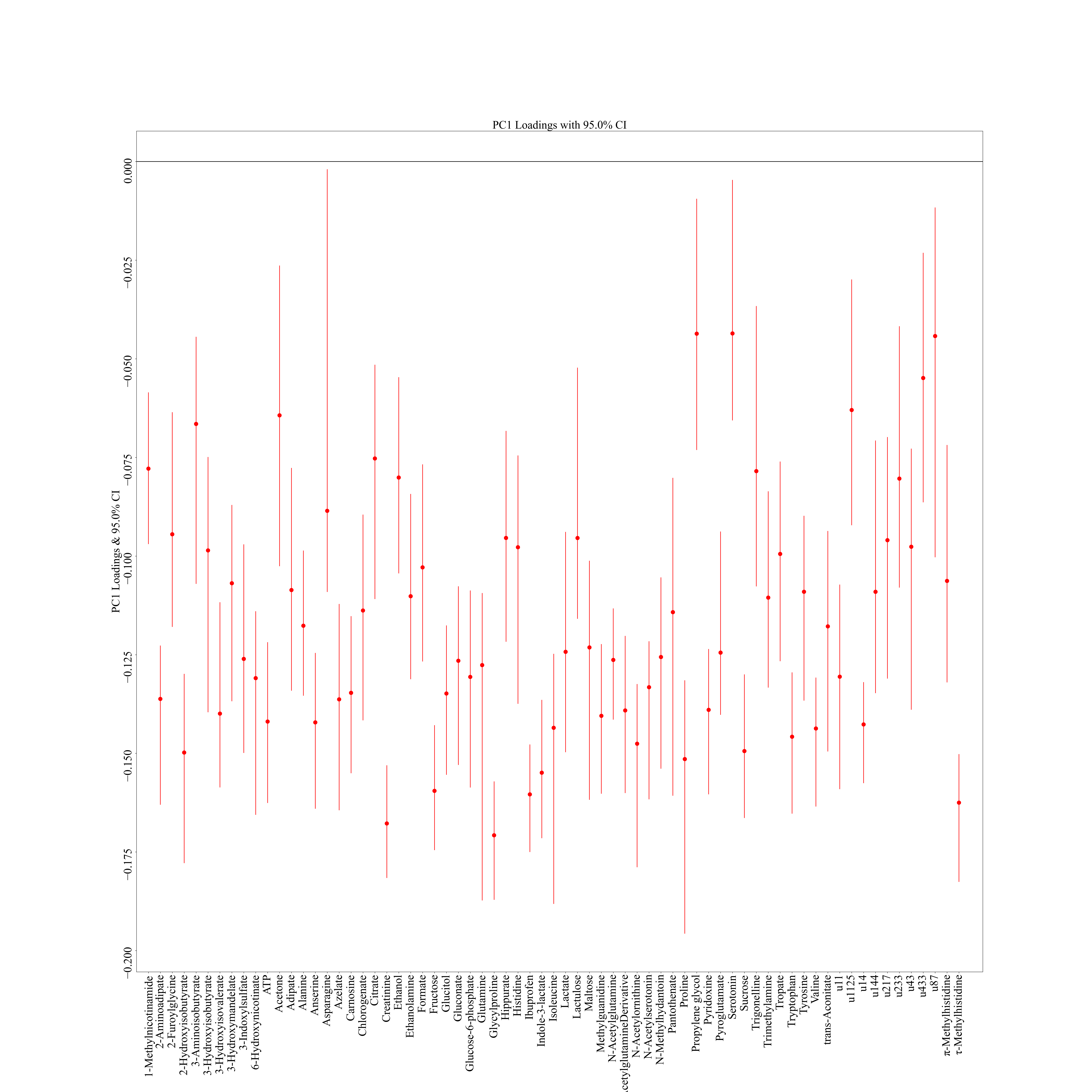
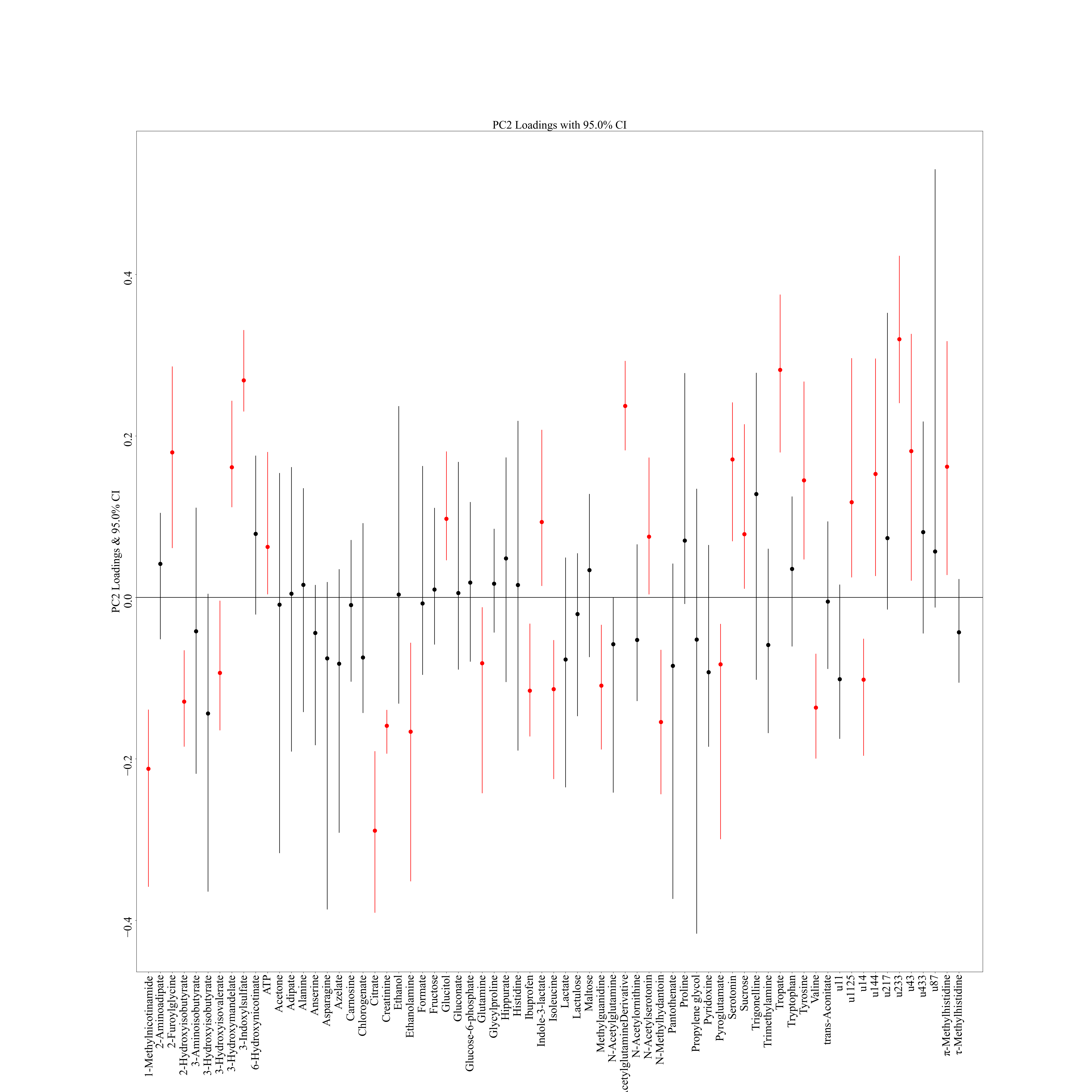
These tutorials step through a workflow to produce a Principal Component Analysis (PCA) with loadings plot, and also PCA lollipop plots. PCA is an exploratory multivariate data analysis technique which given a set of transformed and scaled observable variables can explain the degree of variance across a group of samples through their principal components. PCA is interpreted through the identification of any groups of samples which may cluster within any of the given axes along the principal components, indicating correlation within the groups, but does not indicate to which degree they are correlated. The included loadings plot indicates which features contribute variance to which group of samples, and depending where they lay in relation to the axes indicate the relative direction of metabolite regulation. A PCA lollipop plot with bootstrapped confidence intervals is also included to indicate which features contribute to each component, and which are statistically significant.
Launch tutorial 1.1 by clicking the "Launch Binder" icon:
Launch tutorial 2.1 by clicking the "Launch Binder" icon:
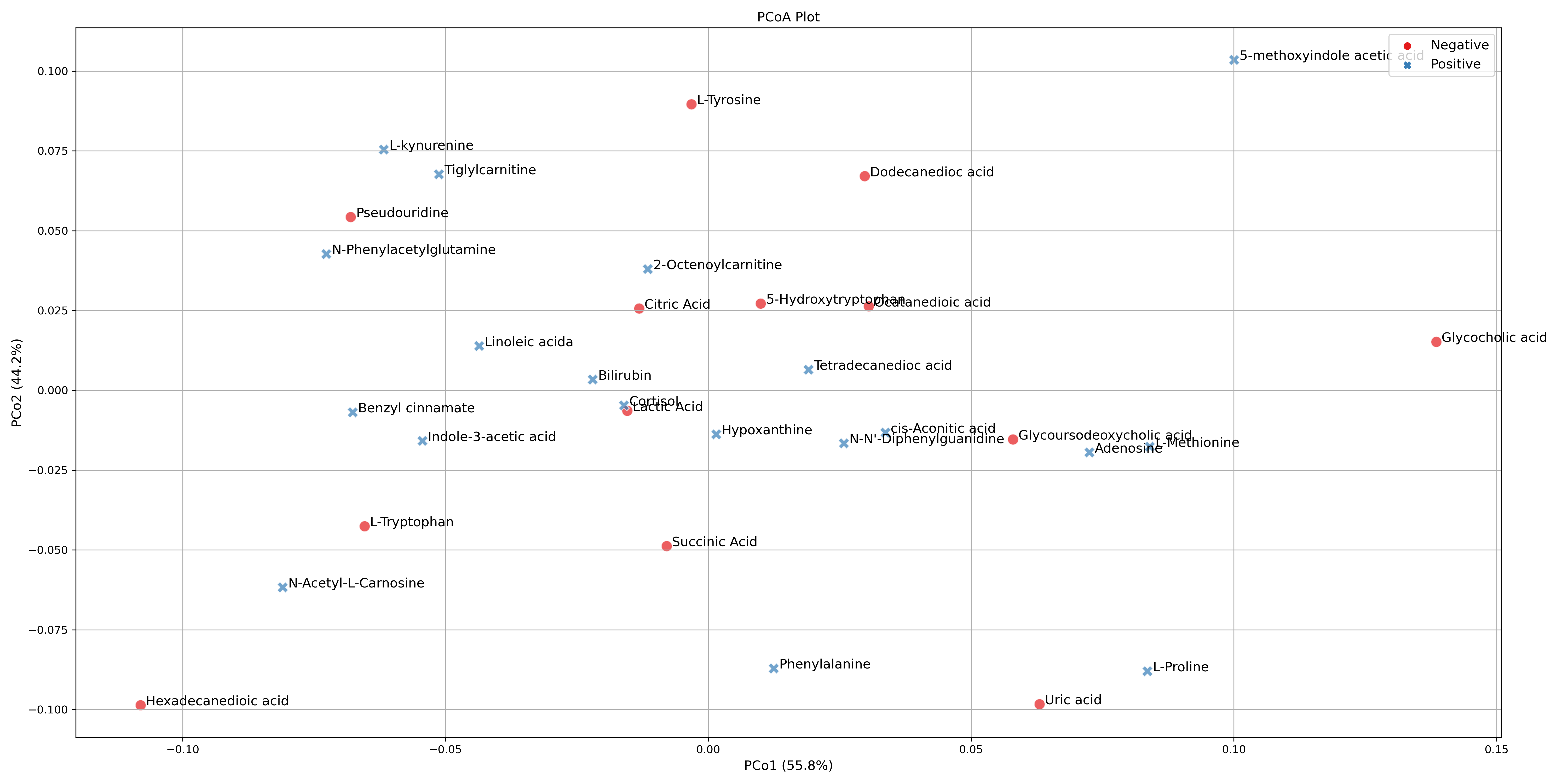
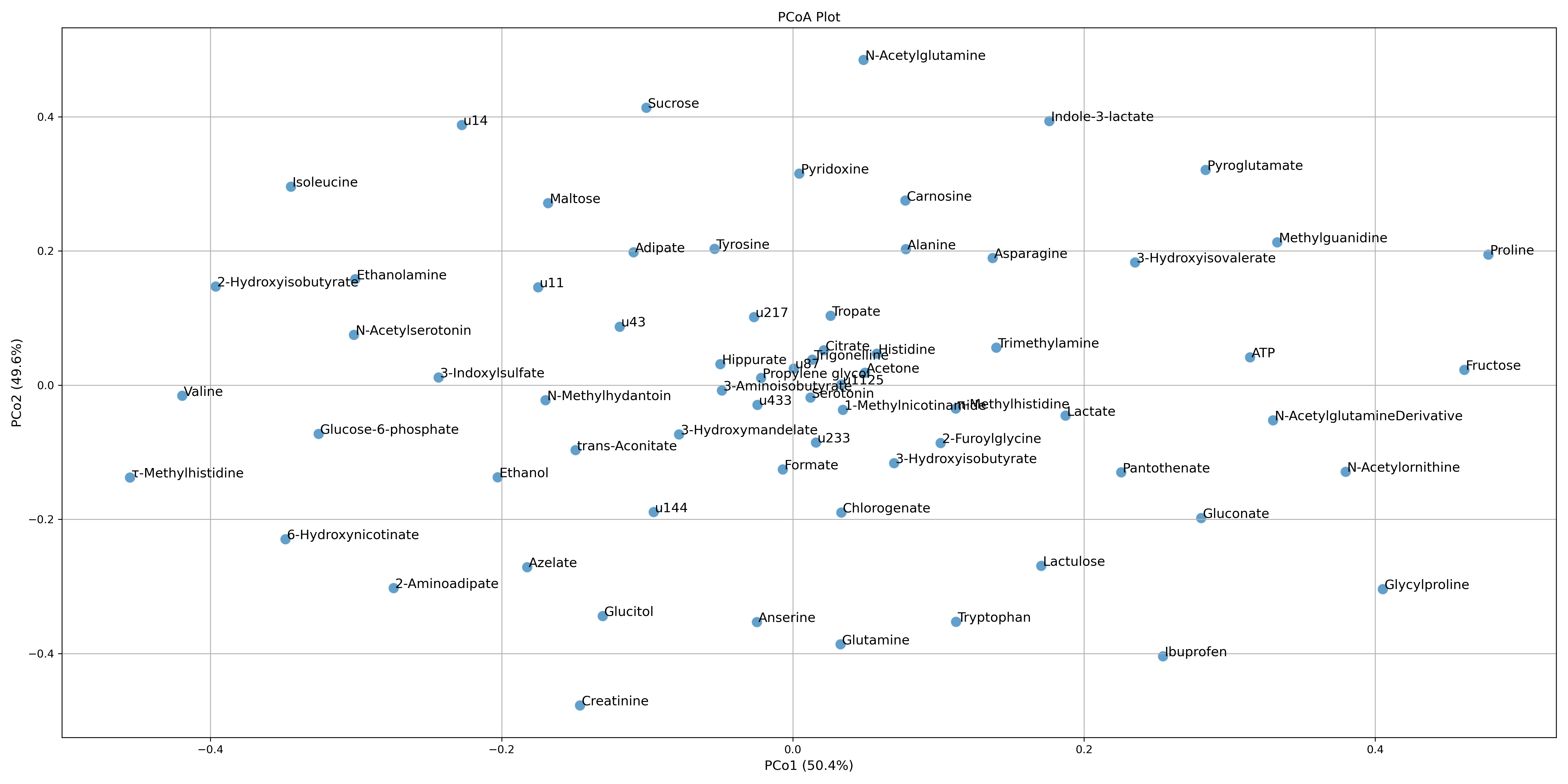
These tutorials step through a workflow to produce a Principal Coordinate Analysis (PCoA) plot. PCoA, also known as Classical Multidimensional Scaling (CMDS), is another multivariate data analysis or ordination method similar to PCA, however it takes a similarity matrix and instead of computing the maximal variance like with PCA, it computes the maximal linear correlation between similarities in the similarity matrix by way of PCA, and projects them to a lower dimensional space.
Launch tutorial 1.2 by clicking the "Launch Binder" icon:
Launch tutorial 2.2 by clicking the "Launch Binder" icon:
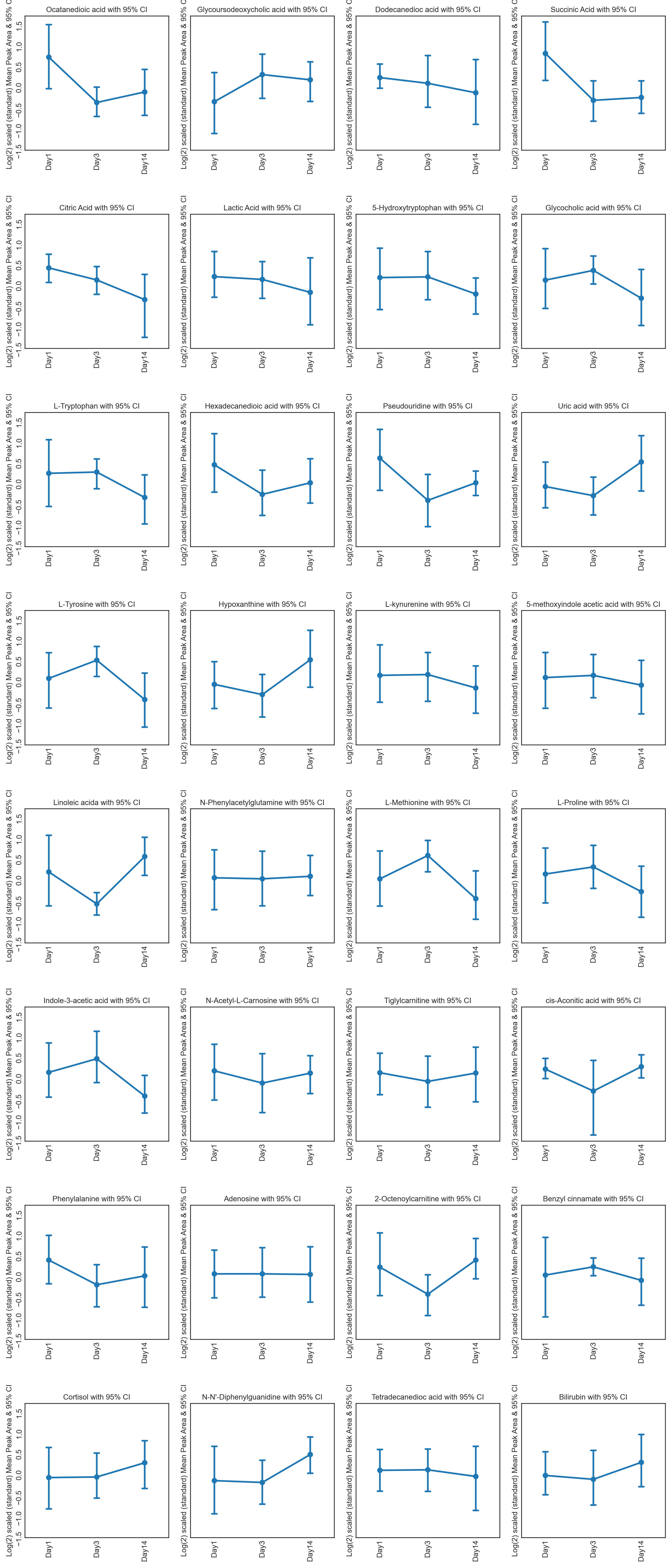
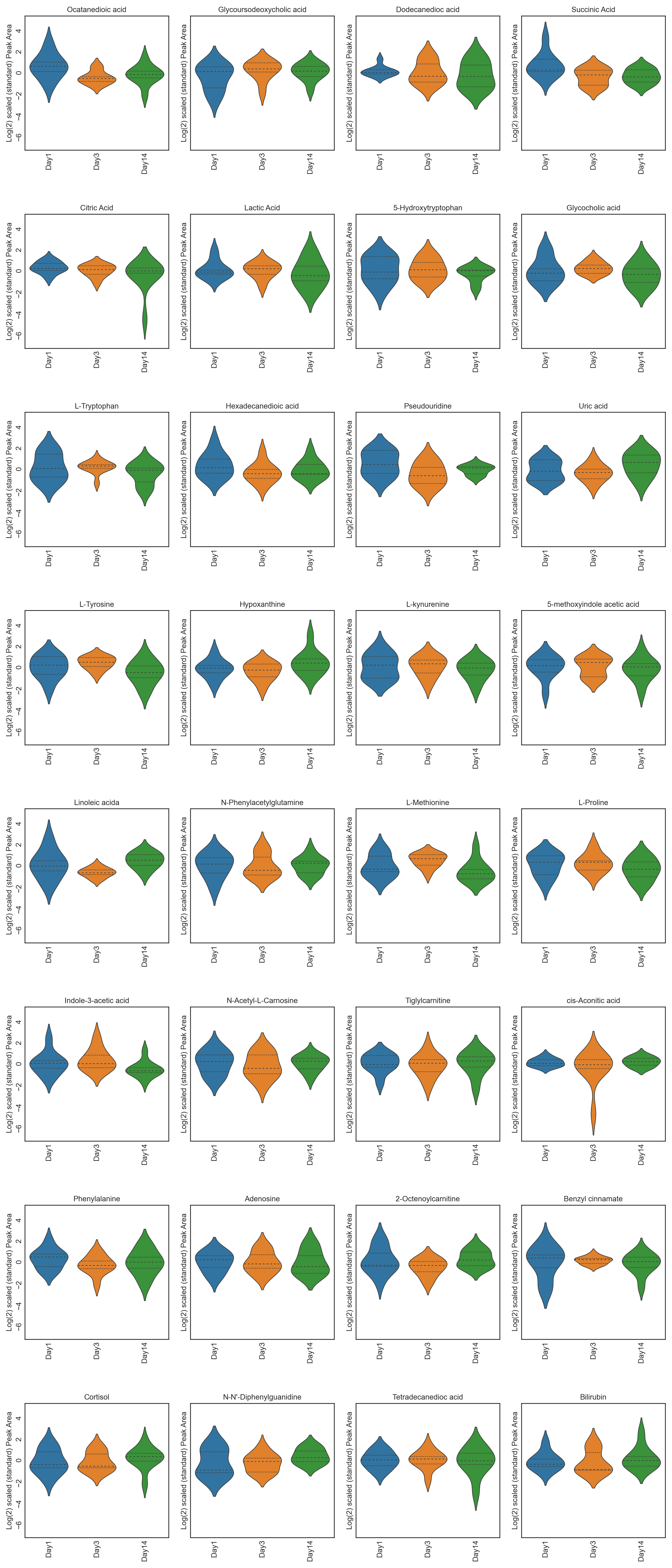
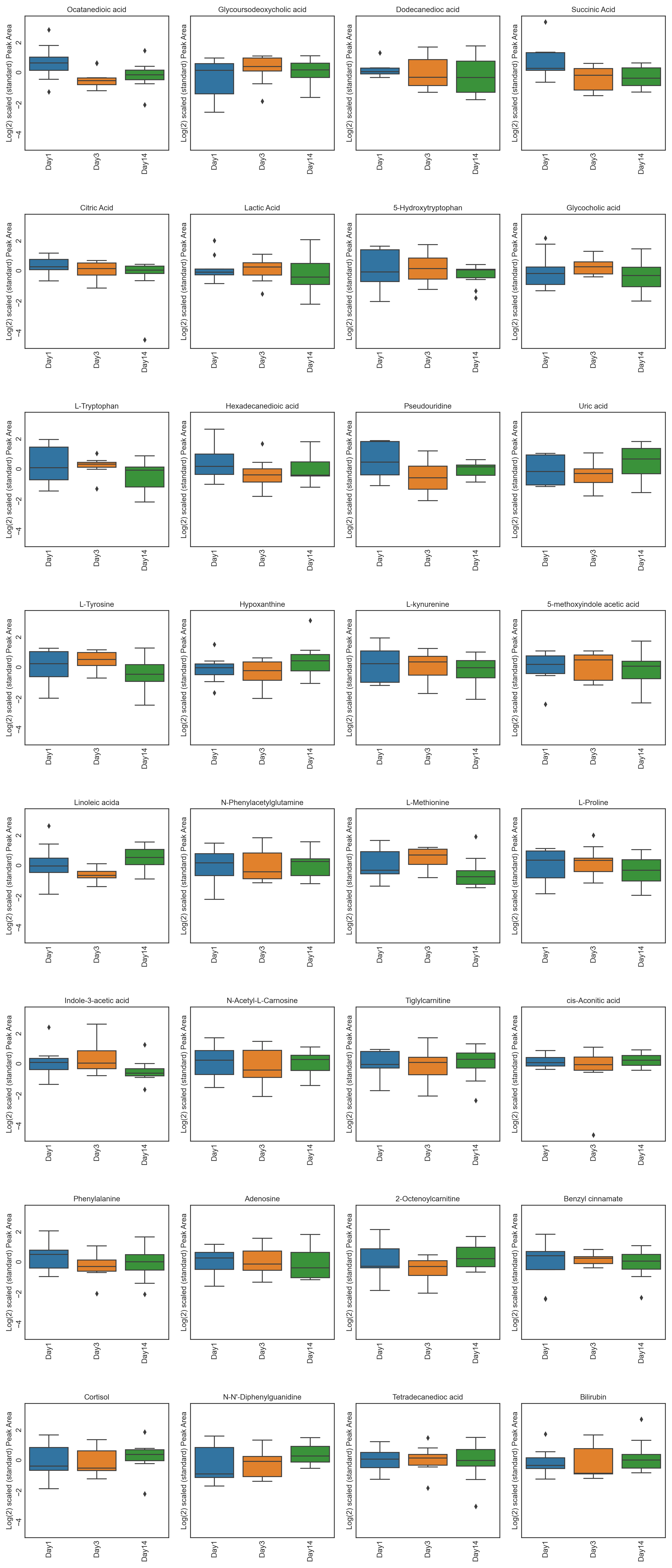
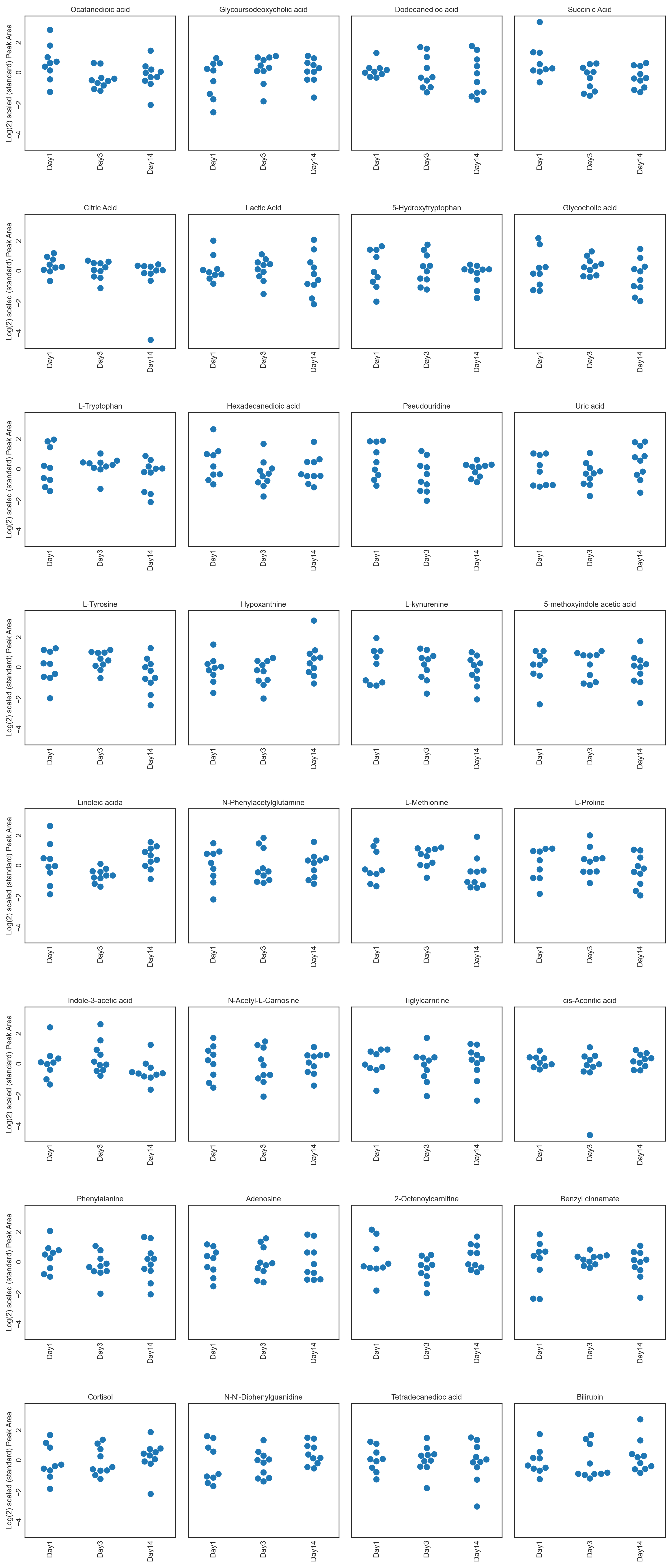
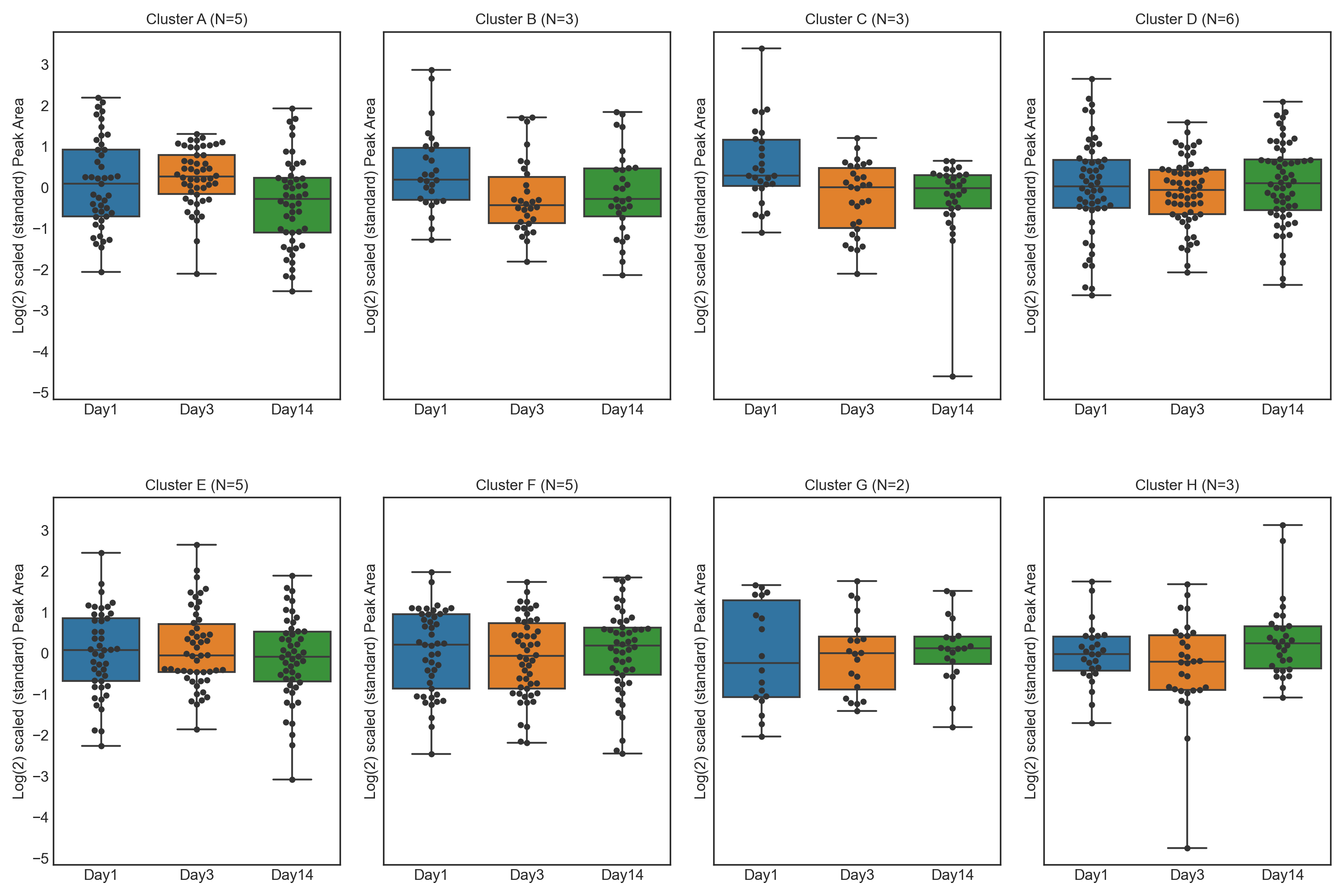
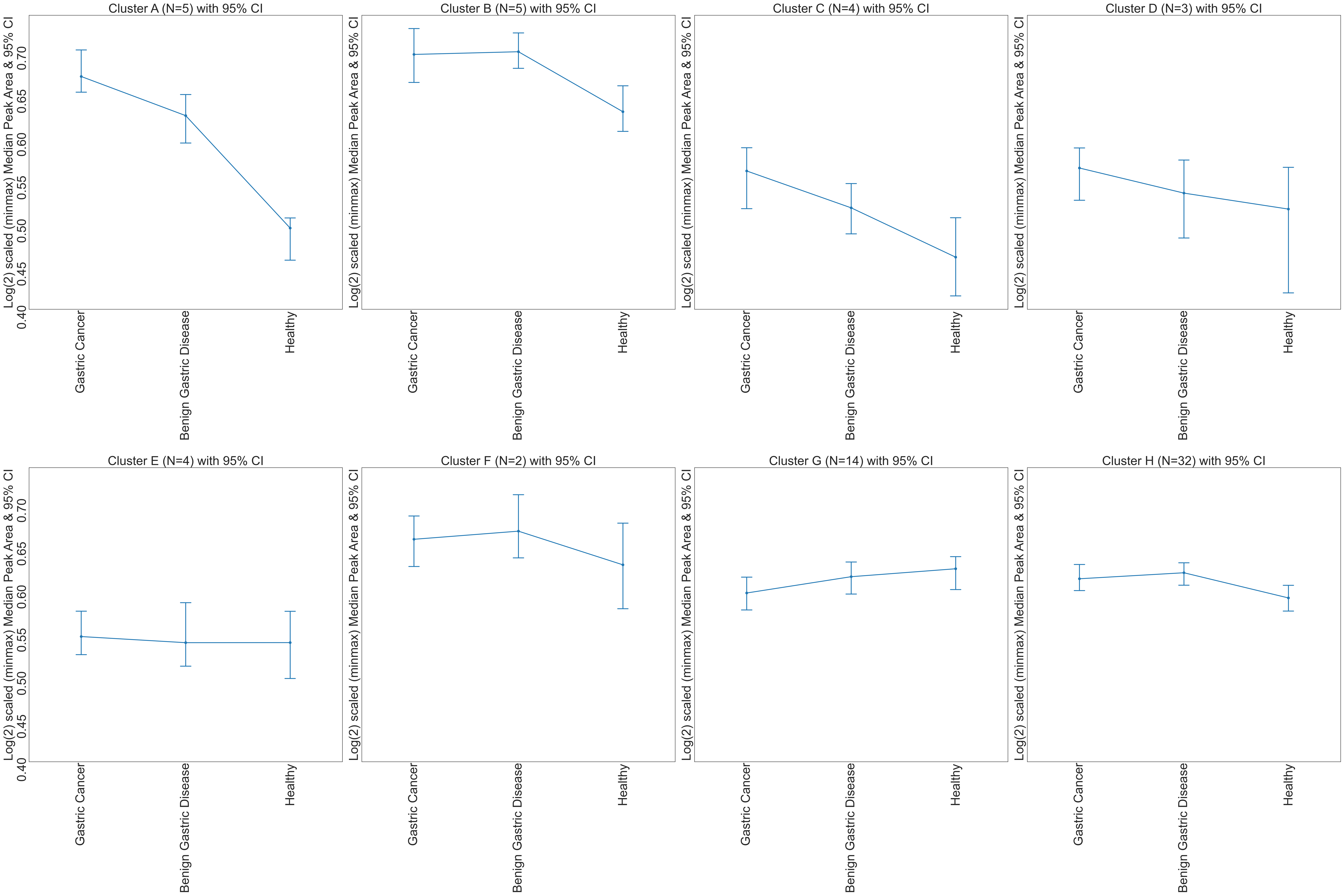
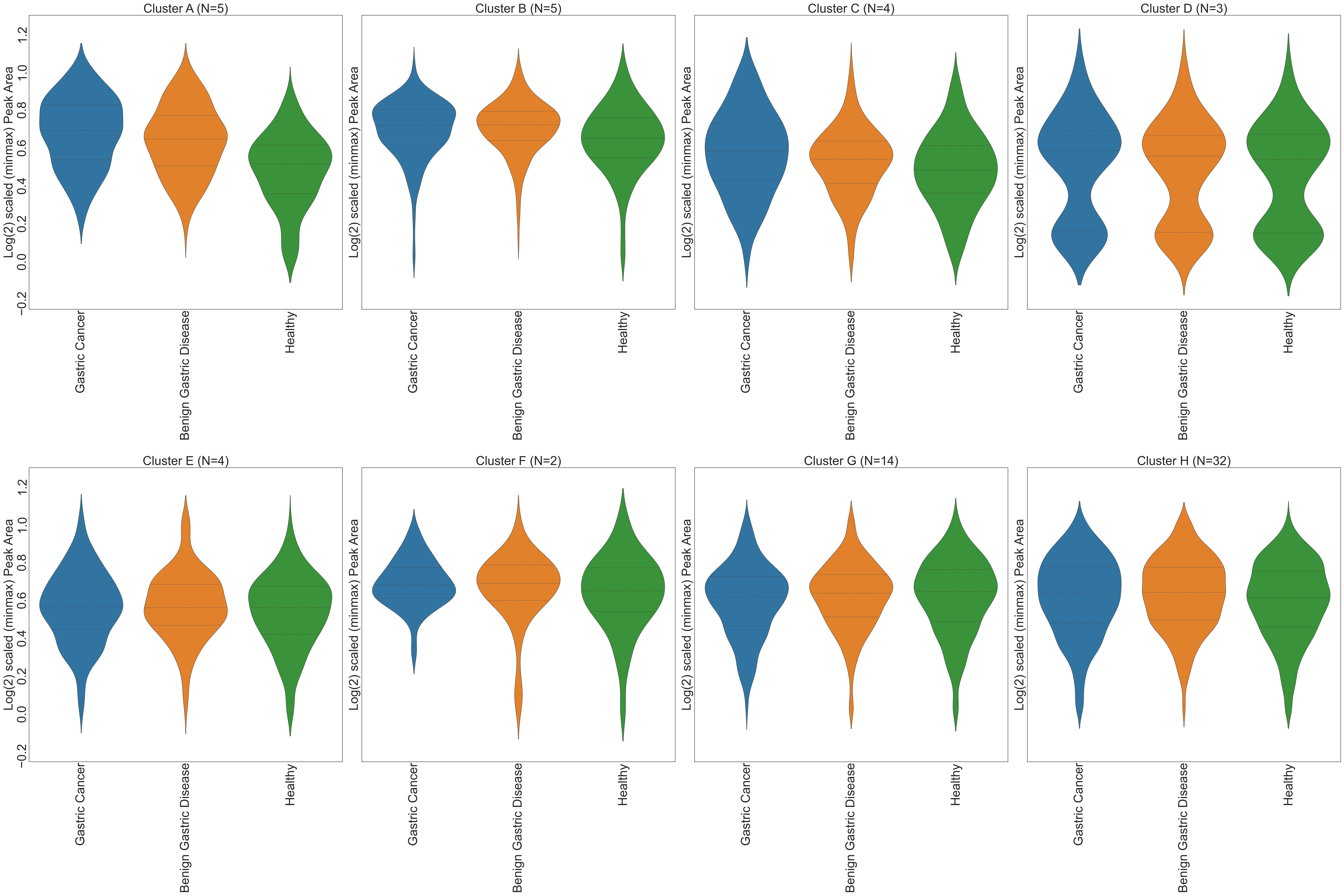
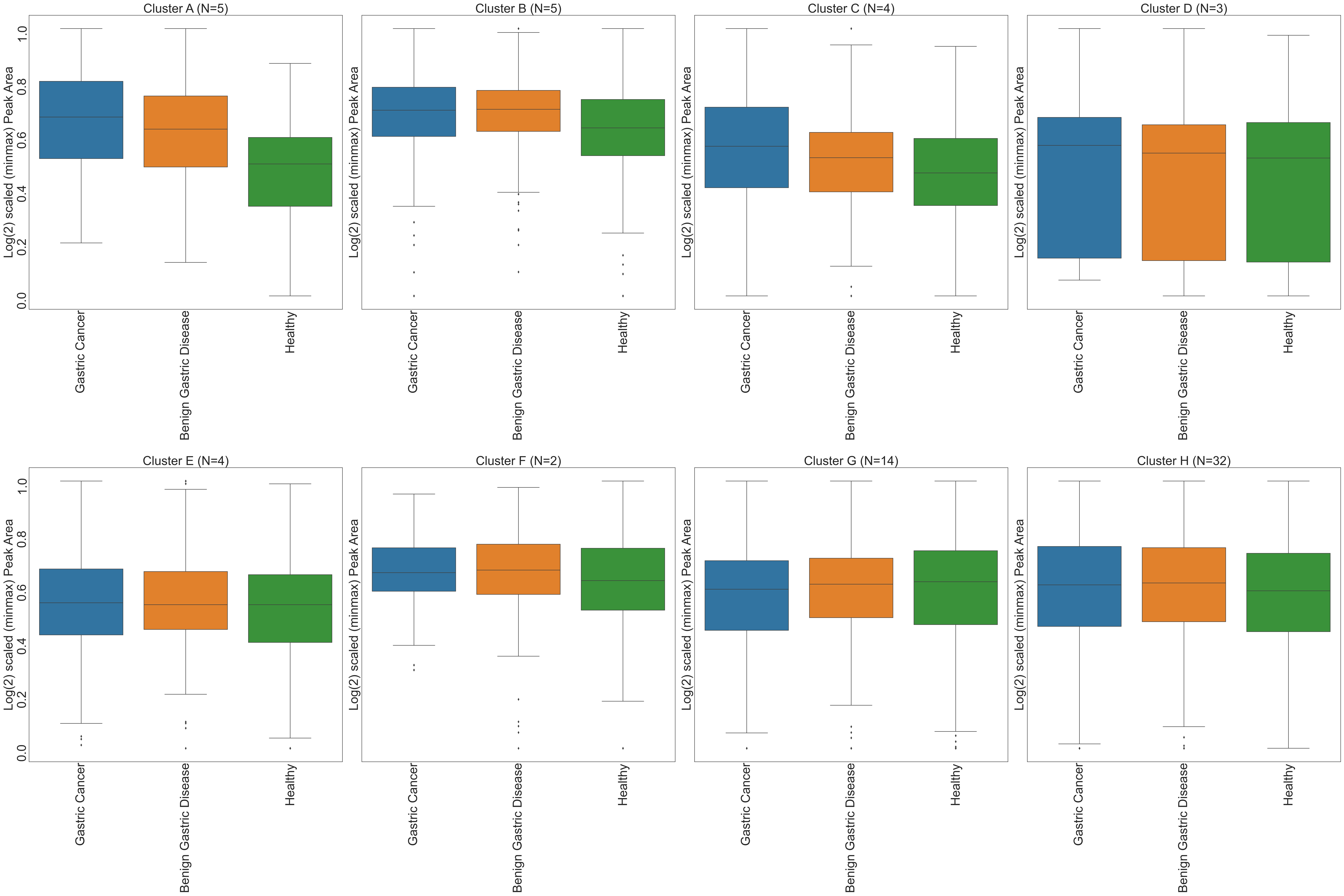
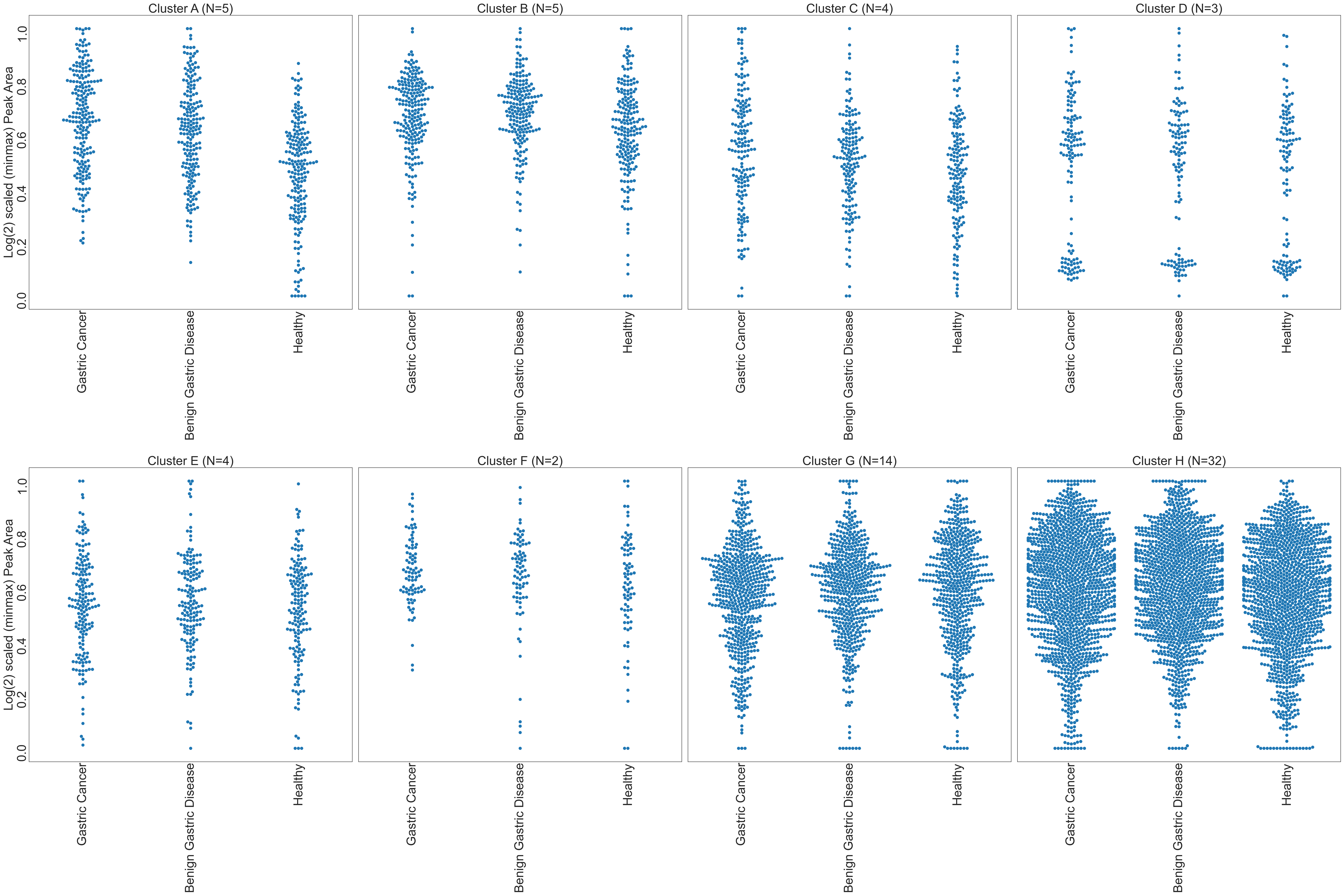
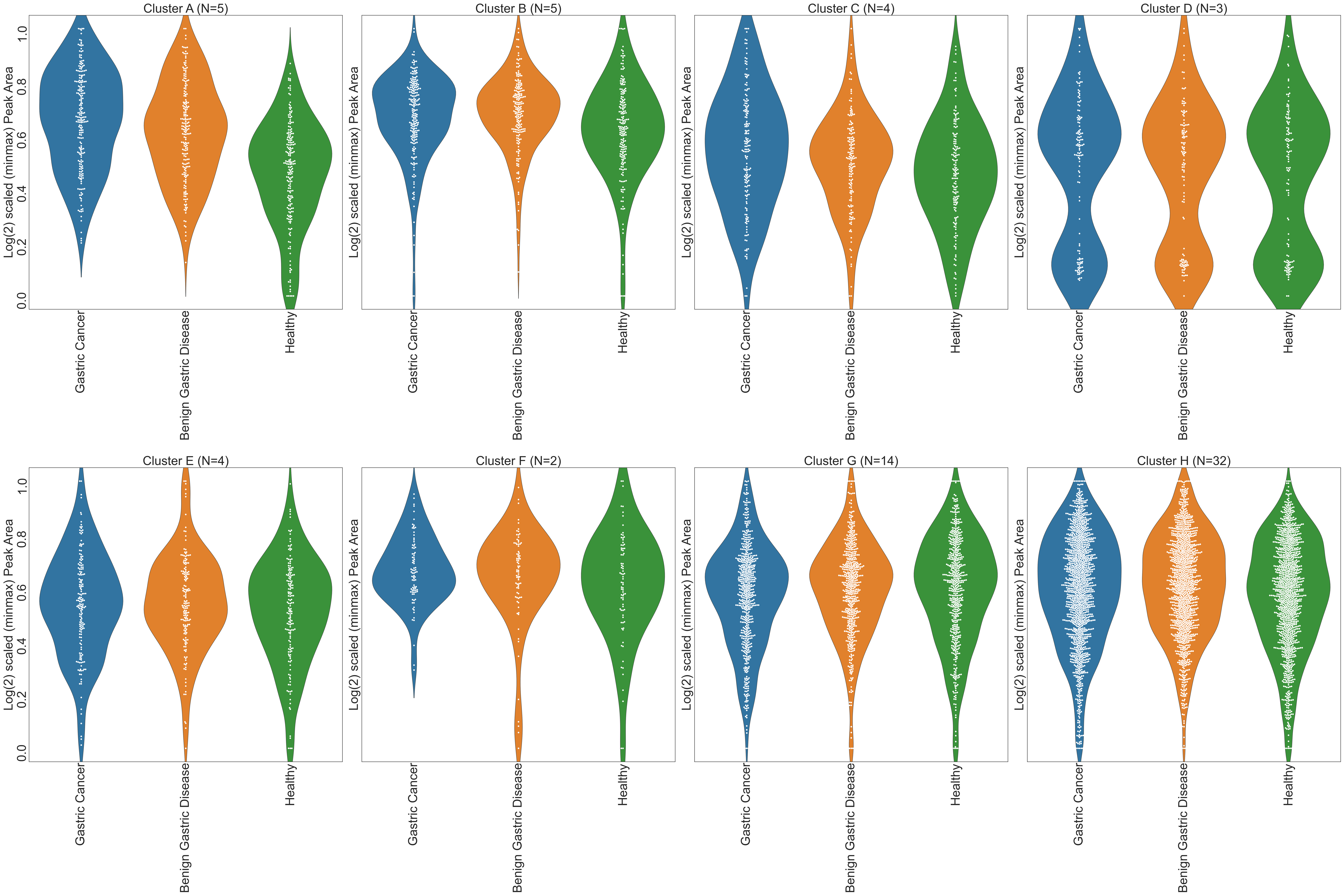
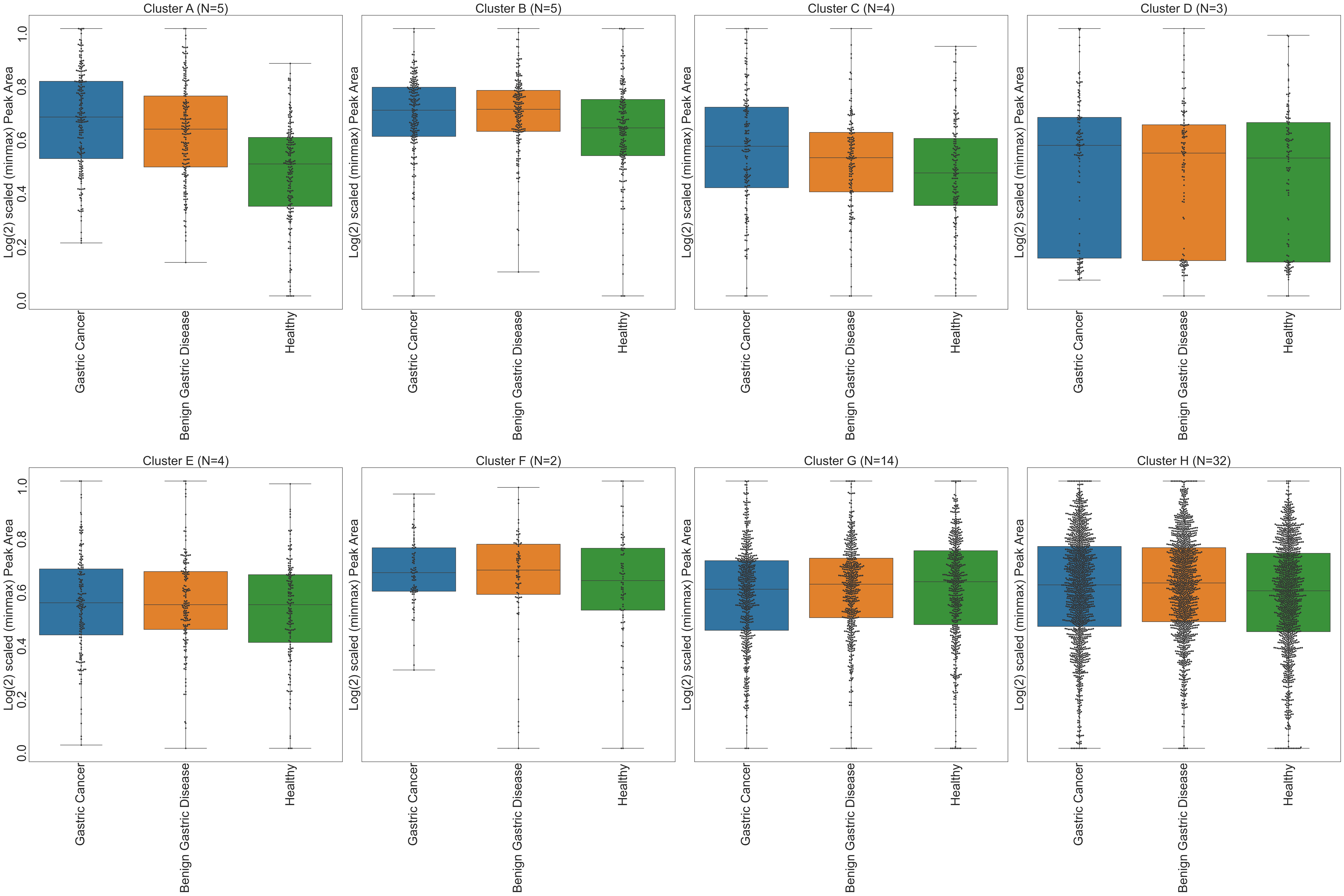
These tutorials step through a workflow to produce different feature plots with the aim of comparing trajectories of features and depending on the type of feature plot, the distribution of each feature can be visualised and compared, providing a way to identify any features which may contain outliers or large differences in magnitude (concentration/expression level), between various groups/classes in the data.
Launch tutorial 1.3 by clicking the "Launch Binder" icon:
Launch tutorial 2.3 by clicking the "Launch Binder" icon:
These tutorials step through a workflow to produce a Hierarchical Clustered Heatmap (HCH). HCH is an exploratory data analysis technique which implements a two-way Hierarchical cluster analysis (HCA) (e.g. clustering samples and features (e.g. metabolites)), and provides dendrograms and heatmaps to visualise the clustered data. HCA is a method by which the distance or correlation between individuals is measured, and then grouped together based on the similarities or dissimilarities between the distances or correlations of the individuals. The clustering itself is based on linkage clustering and determines the overall structure of the clusters. HCH is interpreted through visually identify the different clusters in the heatmap as groups of different colour representations of the clustering score, with the highest score representing one colour choice, and the lowest score representing the other colour choice, and everything in between being lighter or darker shade variants. The different clustered colours in the heatmap are also represented in the dendrograms for each axis as clustered branches.
Launch tutorial 1.4 by clicking the "Launch Binder" icon:
Launch tutorial 2.4 by clicking the "Launch Binder" icon:
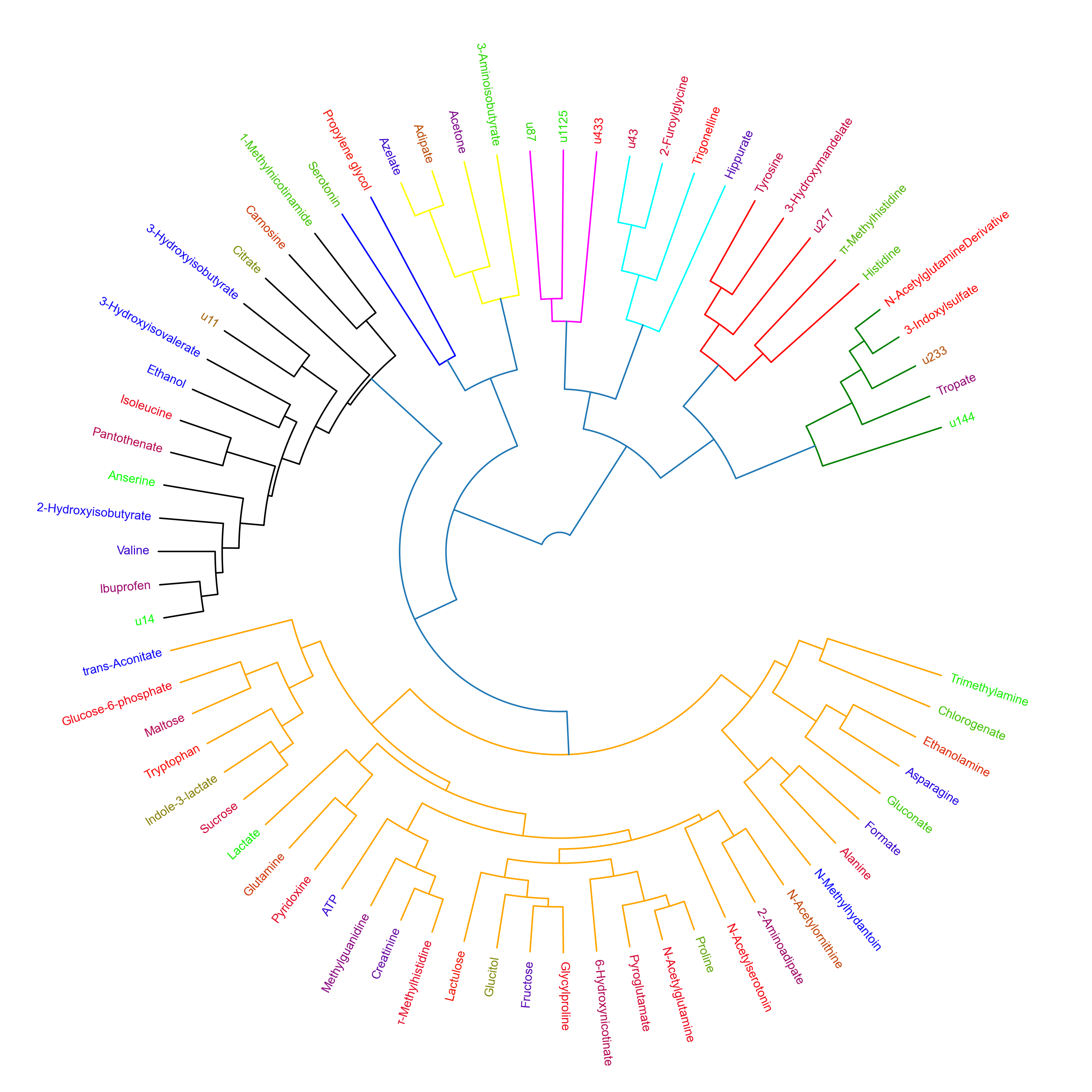
These tutorials step through a workflow to produce a Polar dendrogram. Polar dendrograms are simply a circular form of a dendrogram based on the hierarchical structure of the similarity or dissimilarities from HCA. Polar dendrograms are interpreted by visually identifying the different clustered branches containing closely associated features (e.g. metabolites), the same way with dendrograms in HCH, however now following a circular format. The aggregated peak area of each cluster is then plotted to identify the overall trajectories of features between different groups/classes with the aim of elucidating their biological context.
Launch tutorial 1.5 by clicking the "Launch Binder" icon:
Launch tutorial 2.5 by clicking the "Launch Binder" icon:
These tutorials will step through a workflow to produce a single-block and multi-block spring-embedded network (SEN) plot. SEN plots provide a means to represent how different features are associated with each other, as well their association between different groups/classes, as an undirected graph where each node represents a feature (e.g. metabolite) and the spring constant of each edge between nodes represents some association value such as correlation coefficient. When the tension on the graph is allowed to relax and put in a low energy configuration, the springs push the nodes to their furthest allowed distance from each other in the graph layout. Where there are edges with higher values the spring tightens bringing nodes closer together and where there are lower values the spring loosens and stretches. SEN plots form a type of multivariate cluster analysis, where nodes clustered together are more positively associated together and nodes further apart are considered more negatively associated. The inclusion of univariate and multivariate statistical analysis outputs in the form of pvalues, mean/median fold changes, and principal component loadings, and any other type of statistical outputs can provide another layer of information by illustrating such information as different sizes or colours of nodes. The colours of the nodes can also be associated with other variables such as correlation to drug concentrations, positive/negative fold change, classes of features (e.g. metabolites) or other data types, and the colours of the edges can also be used to represent the positive/negative value of the correlations. Statistical analysis is included in this tutorial to provide a means to filter outliers, remove data with missing values, determine the normality of the data, and to add any further univariate and multivariate statistics to the network plot. Generation of the spring-embedded network is not limited to interpreting matrices of correlation coefficients, but can take any values as long as the matrix contains features on the x-axis and groups/categories/samples on the y-axis. The number of counts of features assigned to different categories can also be used, for example different genomes on the x-axis vs identified protein family domains on the y-axis or using an every day example the number of times individuals visited destinations. All this data can be visualised as a network to identify patterns and previously un-identified associations.
Launch tutorial 1.6 by clicking the "Launch Binder" icon:
Launch tutorial 2.6 by clicking the "Launch Binder" icon:
<iframe width="100%" height="1200" src="springNetwork_negative_single-block_altitude_study.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_single-block_altitude_study_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_negative_single-block_altitude_study_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_multi-block_altitude_study.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_negative_multi-block_altitude_study.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_multi-block_altitude_study_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_negative_multi-block_altitude_study_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_single-block_gastric_cancer.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_single-block_gastric_cancer_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_multi-block_gastric_cancer.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_negative_multi-block_gastric_cancer.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_positive_multi-block_gastric_cancer_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1200" src="springNetwork_negative_multi-block_gastric_cancer_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
These tutorials step through a workflow to produce a multi-block Hierarchical edge bundle (HEB) plot. HEB plots are another means to visualise associations among features and between different groups/classes, in a circular hierarchical tree structure, with nodes on the outside, optionally separated into groups/classes and edges passing through the circle following a bundled curve until they connect to other nodes. The edges represent an association value such as a correlation coefficient and can be coloured accordingly based on the sign of the association value or correlation (e.g. positively vs negatively correlated). Univariate and multivariate statistics can be embedded in the visualisation. Statistical analysis is included in this tutorial to provide a means to filter outliers, remove data with missing values, determine the normality of the data, and to add any further univariate and multivariate statistics to the hierarchical edge bundle.
Launch tutorial 1.7 by clicking the "Launch Binder" icon:
Launch tutorial 2.7 by clicking the "Launch Binder" icon:
<iframe width="100%" height="900" src="hEdgeBundle_multi-block_altitude_study_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="1400" src="hEdgeBundle_multi-block_gastric_cancer.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
<iframe width="100%" height="900" src="hEdgeBundle_multi-block_gastric_cancer_dashboard.html" allowfullscreen="allowfullscreen" frameborder="0"></iframe>
To download the notebook (as changes to the notebook are lost when the Binder session end): 1. Return to Jupyter landing page, by choosing "File" then "Open.." 2. Click the checkbox next to each file you wish to download 3. Click the 'Download' button from the top menu
- Go to the Official Anaconda Website and click the 'Download' button.
- Press the 'Download' button under the 'Python 3.7 version' in Bold to download the graphical installer for your OS.
- After the download has finished, open (double-click) the installer to begin installing the Anaconda Distribution
- Follow the prompts on the graphical installer to completely install the Anaconda Distribution (The final page is 'Thanks for installing Anaconda3')
- Open the app called 'Anaconda Navigator' and press the 'launch' button in the 'Jupyter Notebook' box (to open Jupyter Notebook / confirm it was successfuly installed)
- Go to https://github.com/brettChapman/multivis_tutorials
- Press the green 'Clone or Download', and then click 'Download zip'
- Move this downloaded folder to a suitable directory (by default it is is the 'Download' folder)
- Open the Anaconda Navigator Application
- Move from the 'Home' tab to the 'Environment' tab (on the left side)
- Click import (at the bottom), which opens a box called 'Import New Enviroment'
- In 'Import New Enviroment', press the 'folder' icon and navigate to the 'environment.yml' file in the folder downloaded in step 2/3.
- Click Import and wait for the environment to install (note: may take 5-10 minutes)
- Return to the 'Home' tab
- Change 'Application on... base(root)' to 'Application on... multivis_tutorials' (at the top)
- Press the 'Launch' button under the 'Jupyter Notebook' box
- Using the Notebook Dashboard, locate the downloaded folder in step 2/3 and open (double-click) on the 'Tutorial.ipynb'.
- Open Terminal on Linux/MacOS or Command Prompt on Windows
- Enter the following into the console (one line at a time)
git clone https://github.com/brettChapman/multivis_tutorials
cd multivis_tutorials
conda env create -f environment.yml
source activate multivis_tutorials
jupyter notebookNote: if you receive any of the following errors: "fatal: destination path 'multivis_tutorials' already exists and is not an empty directory" and/or "CondaValueError: prefix already exists: /Users/[insert your username]/opt/anaconda3/envs/multivis_tutorials", then you need to delete the 'multivis_tutorials' folder in the specified directories. The directory can be deleted using Terminal / Command Prompt with:
rm -dfr multivis_tutorials
rm -dfr /Users/[insert your username]/opt/anaconda3/envs/multivis_tutorials