This application is intended to provide a user interface for Gromacs (https://www.gromacs.org/).
For now it is implemented in a way that it allows to quickly set up a simulation by creating an input structure based on a PDB (https://rcsb.org), choosing a force field and a water model as well as a simulation box. Afterwards the input structure is solvated with the water model and the charges are neutralised.
The generated structure can then be used as an input to the Gromacs
mdrun application. The user interface provides means to create
configurations for different simulations steps, e.g. minimisation and
different ensemble simulations (NVT, NPT).
In future versions the tool is intended to support more options of
the Gromacs mdp files. And also guide the user more with reasonable
defaults.
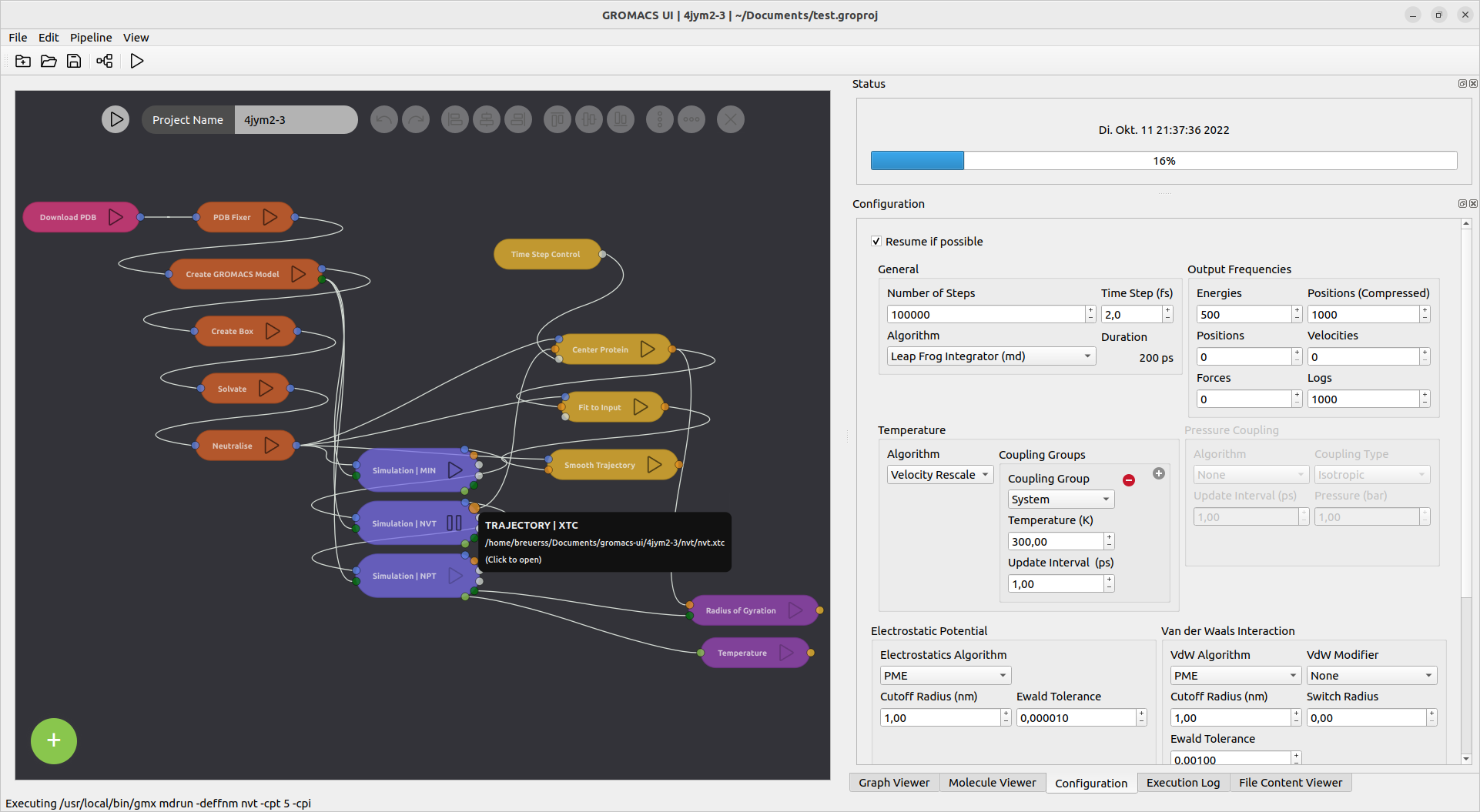
The setup and execution of a simulation can be understood as a pipeline of data passing through modifiying steps. Hence, the new user interface is designed as simulation pipeline configurator. It allows to create a pipeline of nodes connected to each other.
Data can be downloaded from rcsb.org and loaded into the pipeline from hard drive. Several preparation steps modify the input structure and prepare it for the simulation.
Different simulation types can be chained after one another to allow minimisation, equillibration, and production runs in a row.
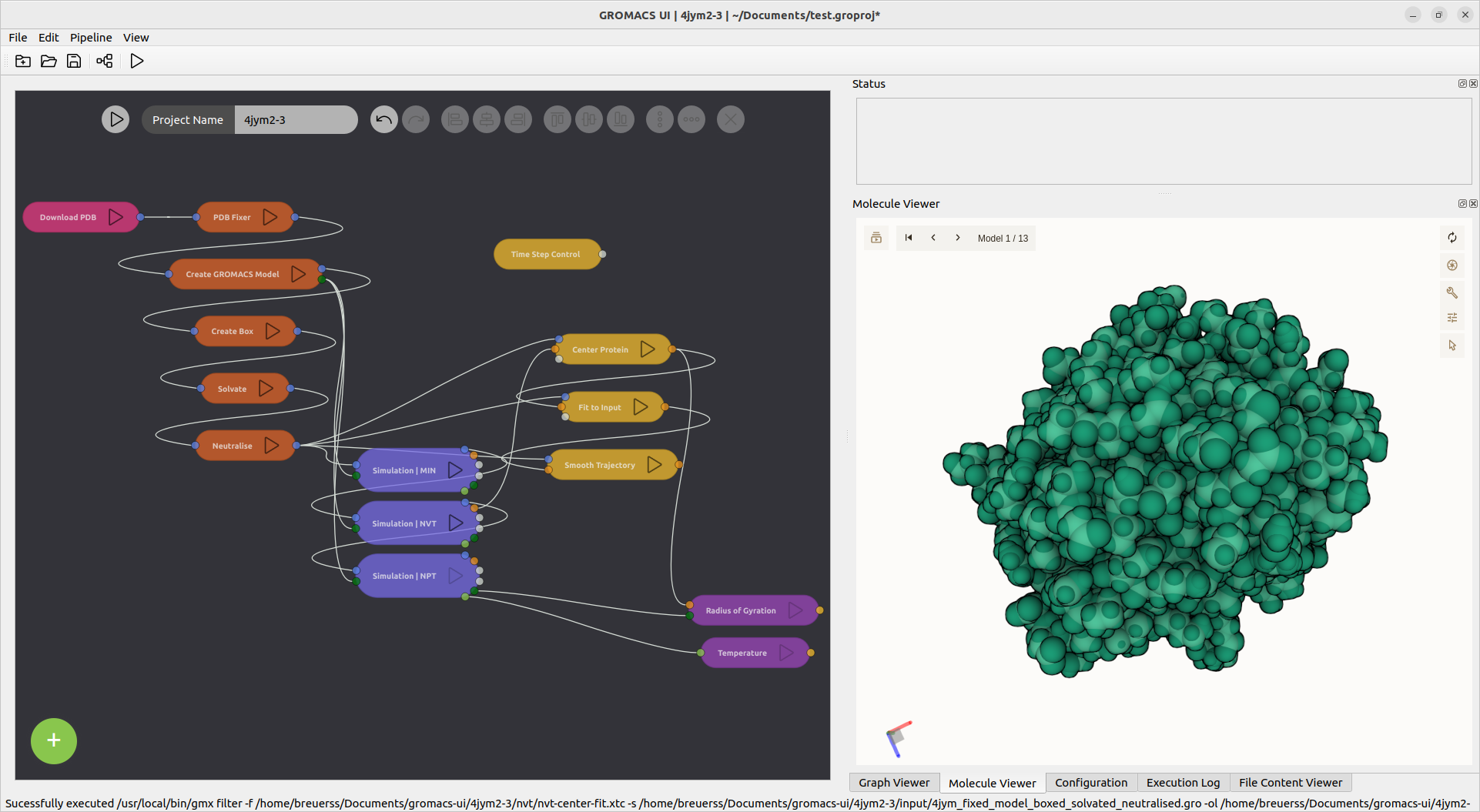
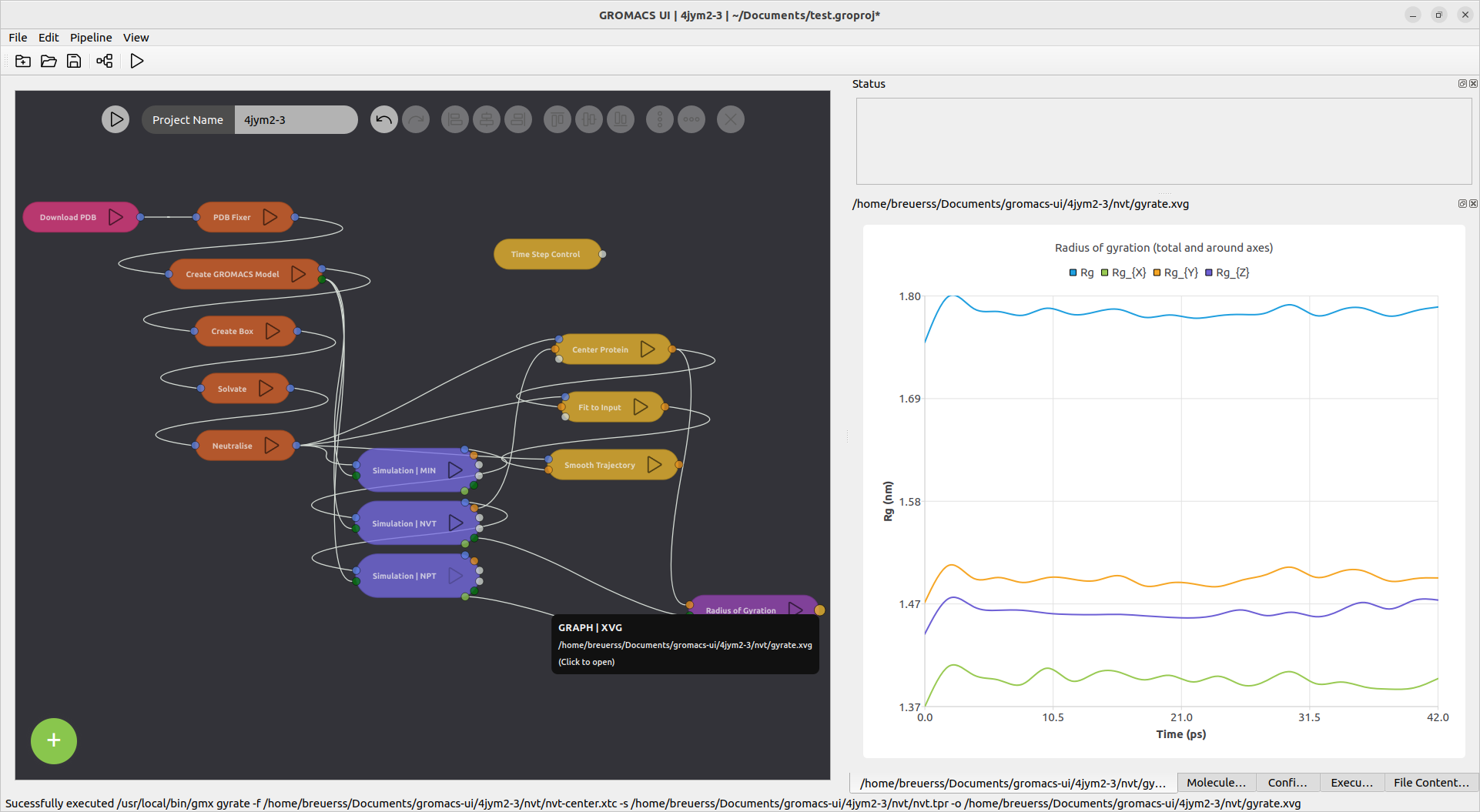
The generated structures and output files can be viewed inside of the application by clicking the respective output ports.
Postprocessing and analysis tools can be added to the chain to clean up the trajectory or extract information from trajrectory or energy files.
The application is based on the Qt framework and tested with version 5.15.3. So the installation of Qt is required to compile the application.
- Qt
- npm (node package manager)
Since this application is a frontend it requires the following tools to be in your applications PATH.
- pdbtools (https://github.com/haddocking/pdb-tools)
- gromacs (https://github.com/gromacs/gromacs)
Please follow the installation instructions provided by those tools.
The molecule and trajectory viewer used inside this tool is the molstar viewer that is also used in the Protein Data Bank (https://rcsb.org).
- molstar (https://github.com/molstar/molstar.git)
For convenience it is added as submodule to this application and needs to compiled in place.
git clone --recurse-submodule git@github.com:breuerss/gromacs-ui.git
cd gromacs-ui
## Compile molstar
cd molstar
npm install && \
npm run build
cd -
## Compile gromacs-ui
mkdir build
cd build
qmake -spec linux-clang .. && \
make -j $(nproc) && \
sudo make install
cd -
After installation the application can be started with
gromacs-ui
- PDB download
- PDB preparation
- Simulation configuration including solvation and neutralisation
- Simulation workflows with minimisation, NVT and NPT
- Simulation configuration, temperature copuling groups
- Trajectory and coordinate visualisation
- Resume simulation
- Simulation progress, tracking minimisation evolution
- Display of logs, mdp configuration, simulation and preparation command output
- Missing analysis views
- Missing trajectory conversions or postprocessing steps, e.g. trajectory smoothing, removal of periodic boundary jumps
- Usage of own prepared structure
- Support of full feature set of gromacs capabilities
- Lingering binary incompatibility of project files between different versions of gromacs-ui, i.e. if new options are added. Project files should be converted to JSON.
- Inconsistency between pdb_delhetatms and pdbfixer.