This repository contains a C++ implementation of Cascade information reconciliation protocol for Quantum Key Distribution (QKD).
This tutorial provides a detailed description of the Cascade information reconciliation protocol and its relation to Quantum Key Distribution.
The C++ code in this repository is modeled after my earlier Python implementation in GitHub repository Cascade-Python, which is documented on ReadTheDocs.
The C++ code is orders of magnitude faster than the Python code. This is because the C++ code was carefully optimized based on the insights from profiling the code. In particular, the decision to cache Shuffle objects made a huge impact on the performance.
The "make data-papers" target in the Python code does 1,000 Cascade iterations per data point and takes more than 5 days of continuous running on an AWS m5.2xlarge instance (120 hours x US$ 0.40 per hour = US$ 48 in compute cost). By contrast, the "make data-papers" target the C++ code does 10,000 Cascade iterations per data point (10x better accuracy) only takes ten hours to complete (US$ 4).
The C++ was more carefully debugged than the Python code. The C++ code has a "make debug" target that produces color-coded representations of the blocks at each step in the algorithm (bit errors are shown in red). This makes it much easier to follow what is going on in the algorithm. Beyond debugging, this is also a good tool for learning the Cascade algorithm.
As a result of the better debugging in the C++ code, the data produced by the C++ is more consistent with the results published in Demystifying the Information Reconciliation Protocol Cascade and Quantum Key Distribution Post Processing - A study on the Information Reconciliation Cascade Protocol.
To install on Ubuntu 18.04:
git clone https://github.com/brunorijsman/cascade-cpp.git
cd cascade-cpp
sudo apt install -y make
make ubuntu-get-dependencies
To build and run unit tests:
make test
To build and run unit tests, and produce code coverage report in coverage/coverage-test.html (on a Mac this also opens the coverage report in the browser):
make test-coverage
To produce the data for all experiments:
make data
To produce the data for specific set of experiments:
# Choose one of these
make data-papers
make data-papers-subset
make data-performance
make data-zero-handling
To produce graphs, you need to have Cascade-Python installed in the $HOME/cascade-python directory.
The produce the graphs for specific set of experiments (this assumes that that the data has already been produced):
make graphs
To produce the graphs for all experiments:
# Choose one of these
make graphs-papers
make graphs-performance
make graphs-zero-handling
To run a small reconciliation with full debug prints:
make debug
Under the hood, when you issue make data-papers the Makefile executes the following command:
bin/run_experiments study/experiments_papers.json --output-dir study/data/papers
The executable run_experiments does the following:
-
It reads an experiments JSON file which describes which experiments to run.
-
It runs the requested experiments in a multi-threaded fashion using all available CPU cores.
-
It writes the results into one or more data JSON files that can later be visualized into graphs.
The executable run_experiments uses the following syntax:
$ run_experiments --help
Usage: run_experiments [options] experiments-file
Options:
--help Display help
-d [ --disable-multi-processing ] Disable multi-processing
-m [ --max-runs ] arg Maximum number of reconciliation runs per
data point
-s [ --seed ] arg Random seed
-o [ --output-directory ] arg Output directory where to store data__*
files
Here is an example of an experiments JSON file (this is experiments_papers.json):
[
{
"independent_variable": "error_rate",
"algorithm": ["original", "biconf", "yanetal", "option3", "option4", "option7", "option8"],
"error_rate": [
{"start": 0.0000, "end": 0.0050, "step_size": 0.00005},
{"start": 0.0050, "end": 0.1100, "step_size": 0.00050}
],
"key_size": [1000, 2000, 10000],
"runs": 10000
},
{
"independent_variable": "key_size",
"algorithm": ["original", "biconf", "yanetal", "option3", "option4", "option7", "option8"],
"error_rate": 0.05,
"key_size": {
"start": 1e3,
"end": 1e5,
"step_factor": 1.05
},
"runs": 10000
},
{
"independent_variable": "key_size",
"algorithm": "original",
"error_rate": [0.01, 0.02],
"key_size": {
"start": 1e3,
"end": 1e5,
"step_factor": 1.05
},
"runs": 10000
}
]
The experiments JSON file consists of a list of records, each describing a single experiment. In this example there are three experiments.
Each experiment has the following arguments.
-
independent_variable: This must beerror_rateorkey_size.-
If it is set to
error_rate, then one separate data file is produced for each requestedkey_size(for exampledata__algorithm=option3;key_size=10000;error_rate=vary) and theerror_rateis varied within each data file. -
If it is set to
key_size, then one separate data file is produced for each requestederror_rate(for exampledata__algorithm=option3;key_size=vary;error_rate=0.05) and thekey_sizeis varied within each data file.
-
-
algorithm: Either a single algorithm or a list of algorithms. Each algorithm is plotted as a line in the graph. The available algorithms areoriginal,biconf,biconf-cascade,biconf-no-complement,yanetal,option3,option4,option7, andoption8. See source filealgorithm.cppfor the detailed parameters for each algorithm. -
error_rate: The error rates to be used in the experiment in the form of a numerical list (see below). -
key_size: The key sizes to be used in the experiment in the form of a numerical list (see below). -
run: The number of reconciliation runs per data point in the results.
A numerical list is one of the following:
-
A list of numbers, for example
[1, 2, 3, 4] -
A linear range of numbers, for example
{"start": 100.0, "end": 200.0, "step": 5.0} -
A log range of numbers, for example
{"start": 10.0, "end": 10000.0, "step_factor": 3.0} -
A list of any of the above.
The executable run_experiments produces a set of data JSON files in the requested output
directory, for example:
$ ls -1 study/data/papers/
data__algorithm=biconf;key_size=10000;error_rate=vary
data__algorithm=biconf;key_size=1000;error_rate=vary
data__algorithm=biconf;key_size=2000;error_rate=vary
data__algorithm=biconf;key_size=vary;error_rate=0.05
...
data__algorithm=yanetal;key_size=2000;error_rate=vary
data__algorithm=yanetal;key_size=vary;error_rate=0.05
Each data JSON file consists of a list of JSON records, one for each data point. Here is an example data point record:
{
"actual_bit_error_rate": {"average": 0.000050, "deviation": 0.000050},
"actual_bit_errors": {"average": 0.502200, "deviation": 0.502200},
"algorithm_name": "biconf",
"ask_parity_bits": {"average": 2828.808000, "deviation": 2828.808000},
"ask_parity_blocks": {"average": 35.360100, "deviation": 35.360100},
"ask_parity_messages": {"average": 37.360100, "deviation": 37.360100},
"biconf_iterations": {"average": 10.000000, "deviation": 10.000000},
"elapsed_process_time": {"average": 0.001794, "deviation": 0.001794},
"elapsed_real_time": {"average": 0.001795, "deviation": 0.001795},
"execution_time": "2020-04-08 20:20:49 UTC",
"infer_parity_blocks": {"average": 6.919900, "deviation": 6.919900},
"key_size": 10000,
"normal_iterations": {"average": 2.000000, "deviation": 2.000000},
"realistic_efficiency": {"average": 510.625974, "deviation": 510.625974},
"reconciliations": 10000,
"remaining_bit_error_rate": {"average": 0.000000, "deviation": 0.000000},
"remaining_bit_errors": {"average": 0.000000, "deviation": 0.000000},
"remaining_frame_error_rate": {"average": 0.000000, "deviation": 0.000000},
"reply_parity_bits": {"average": 35.360100, "deviation": 35.360100},
"requested_bit_error_rate": 0.000050,
"unrealistic_efficiency": {"average": 4.495774, "deviation": 4.495774}
}
We rely on the Python scripts in Cascade-Python to produce graphs from the results stored in the
data file produced by the executable run_experiments.
Under the hood, when you issue make graphs-papers the Makefile activates a Python3 virtual
environment and executes the following Python script:
python $(CASCADE_PYTHON_DIR)/study/make_graphs.py study/graphs_demystifying.json \
--data-dir study/data/papers
The graph JSON file (in this example graphs_demystifying.json) describes which variables
should be plotted in the graph and other attributes that influence the appearance of the graph.
This is an example of a graph file (namely the first graph in graphs_demystifying.json):
[
{
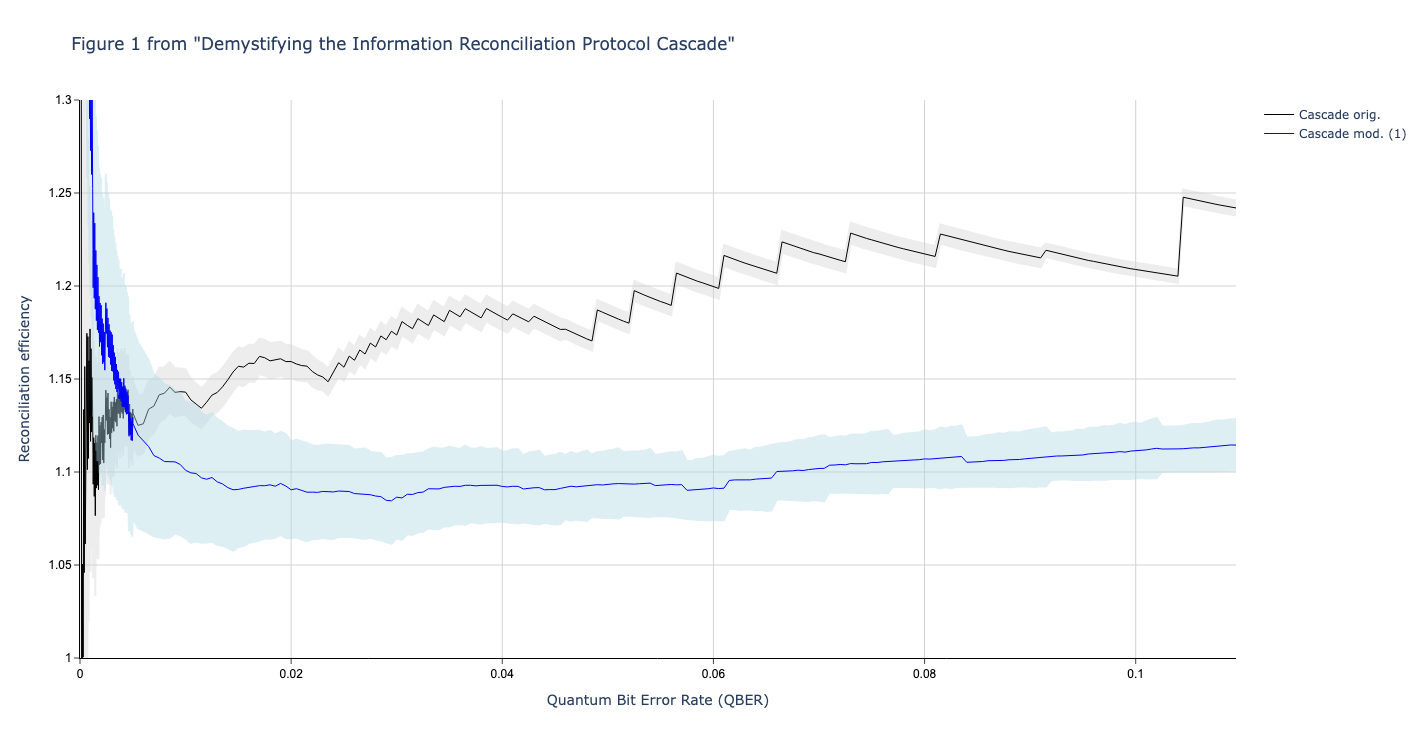
"graph_name": "demystifying_figure_1",
"title": "Figure 1 from \"Demystifying the Information Reconciliation Protocol Cascade\"",
"x_axis": {
"title": "Quantum Bit Error Rate (QBER)",
"variable": "requested_bit_error_rate"
},
"y_axis": {
"title": "Reconciliation efficiency",
"variable": "unrealistic_efficiency",
"range": [1.0, 1.3]
},
"series": [
{
"data_file": "data__algorithm=original;key_size=10000;error_rate=vary",
"legend": "Cascade orig.",
"line_color": "black",
"deviation_color": "lightgray"
},
{
"data_file": "data__algorithm=biconf;key_size=10000;error_rate=vary",
"legend": "Cascade mod. (1)",
"line_color": "blue",
"deviation_color": "lightblue"
}
]
},
...
]
This produces the following graph:
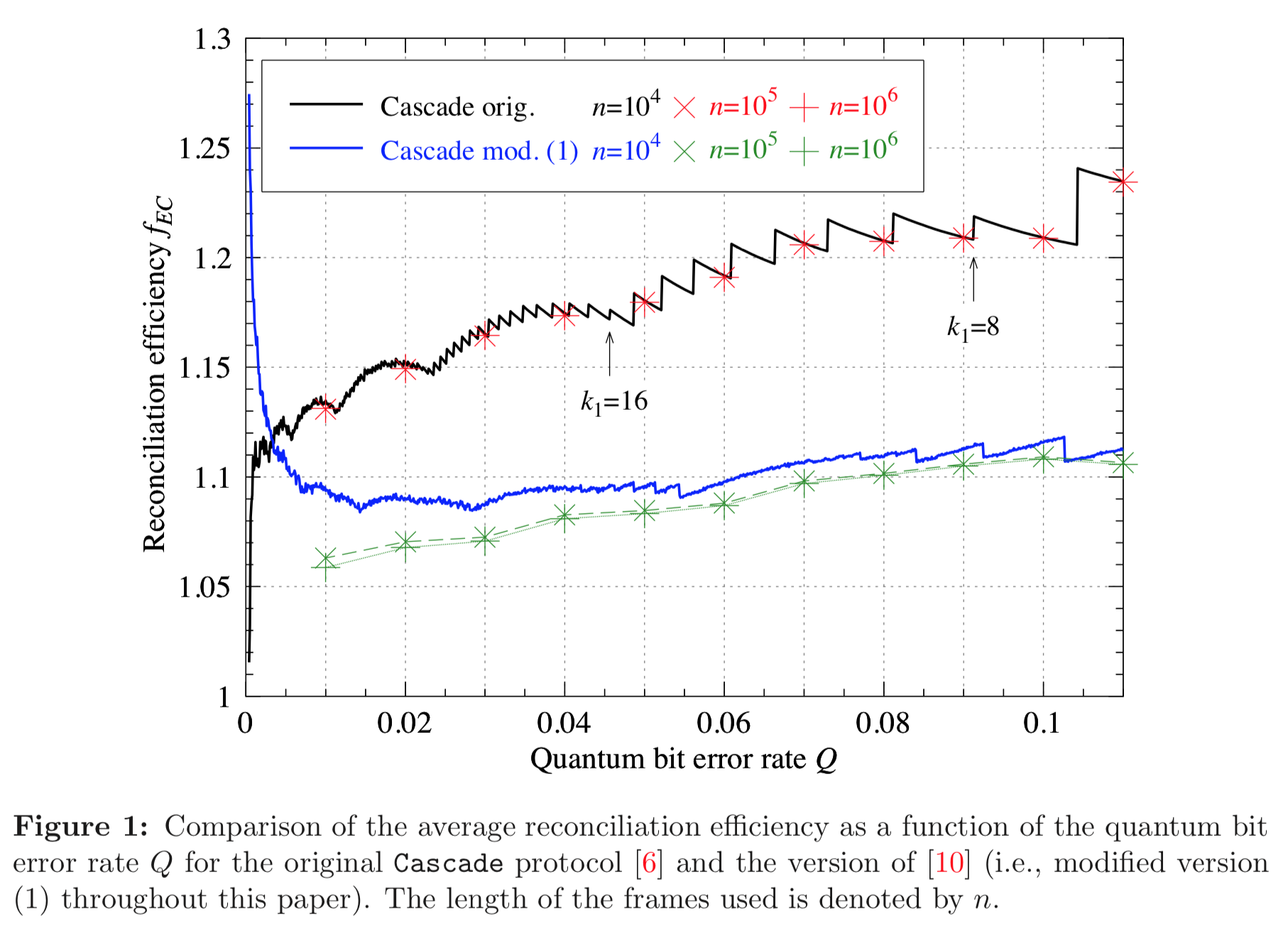
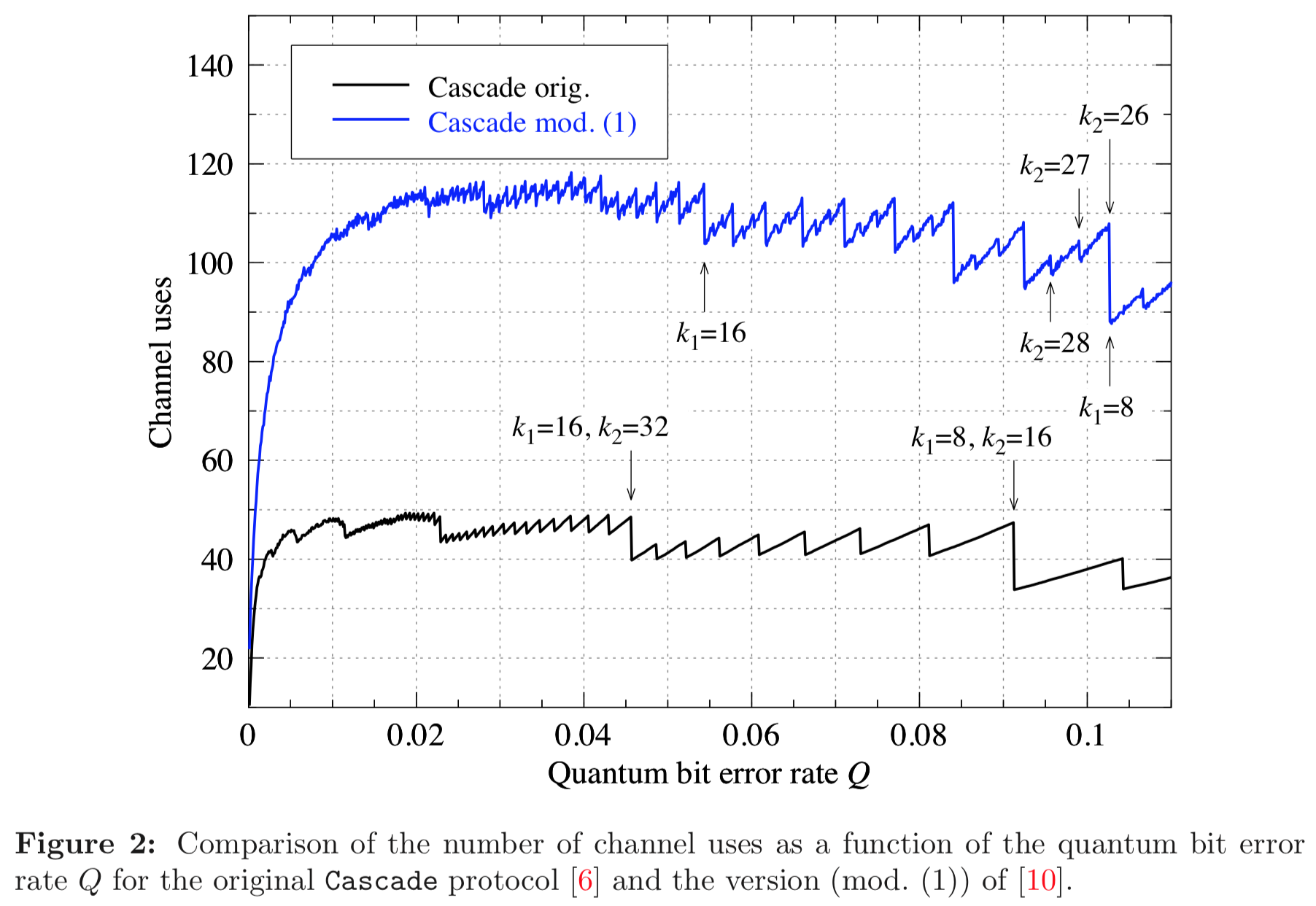
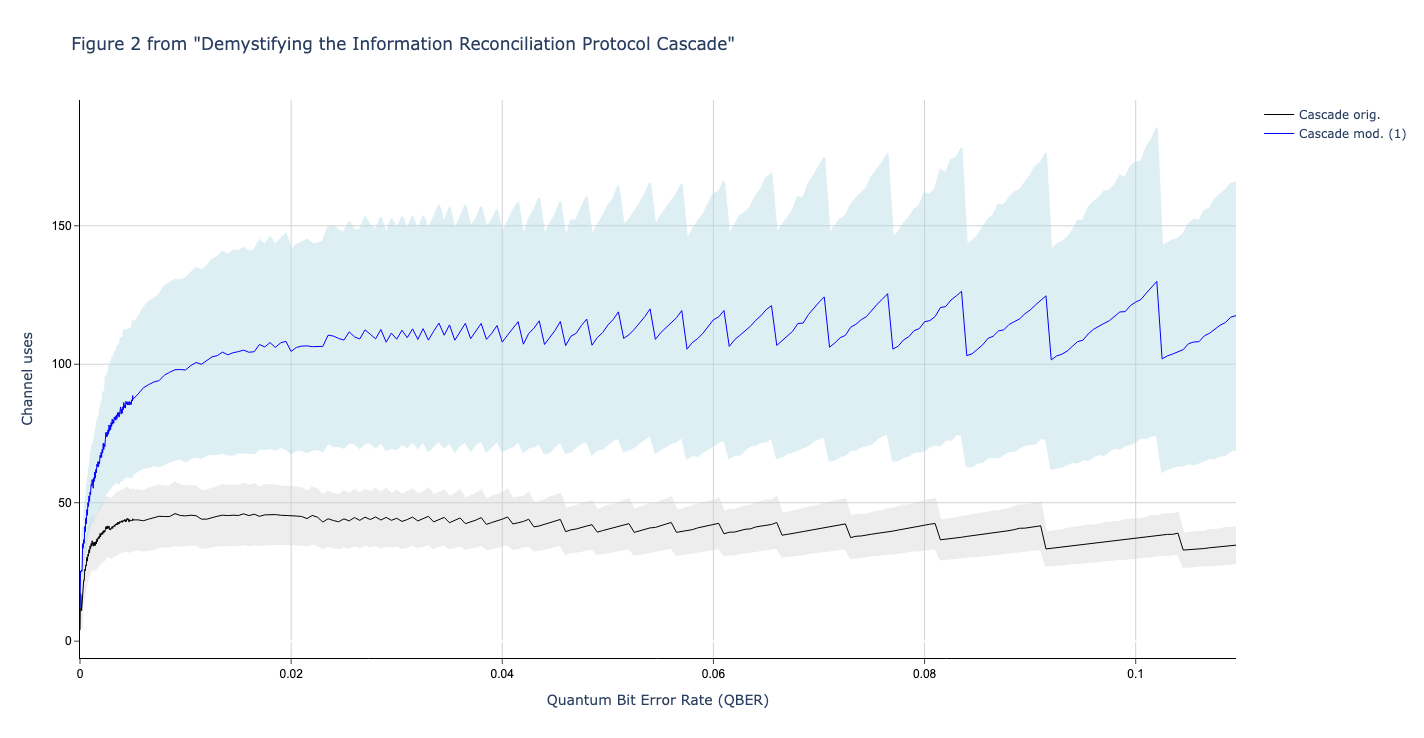
In this section we compare the data produced
We used Github version c4d2194bcae4580f329597677ba2e2538bb53c81 (8 April 2020) of Cascade-CPP to produce the data in these graphs.
In this section we compare the data produced by Cascade-CPP with the results reported in:
Demystifying the Information Reconciliation Protocol Cascade.
Jesus Martinez-Mateo, Christoph Pacher, Momtchil Peev, Alex Ciurana, and Vicente Martin.
arXiv:1407.3257 [quant-ph], Jul 2014.
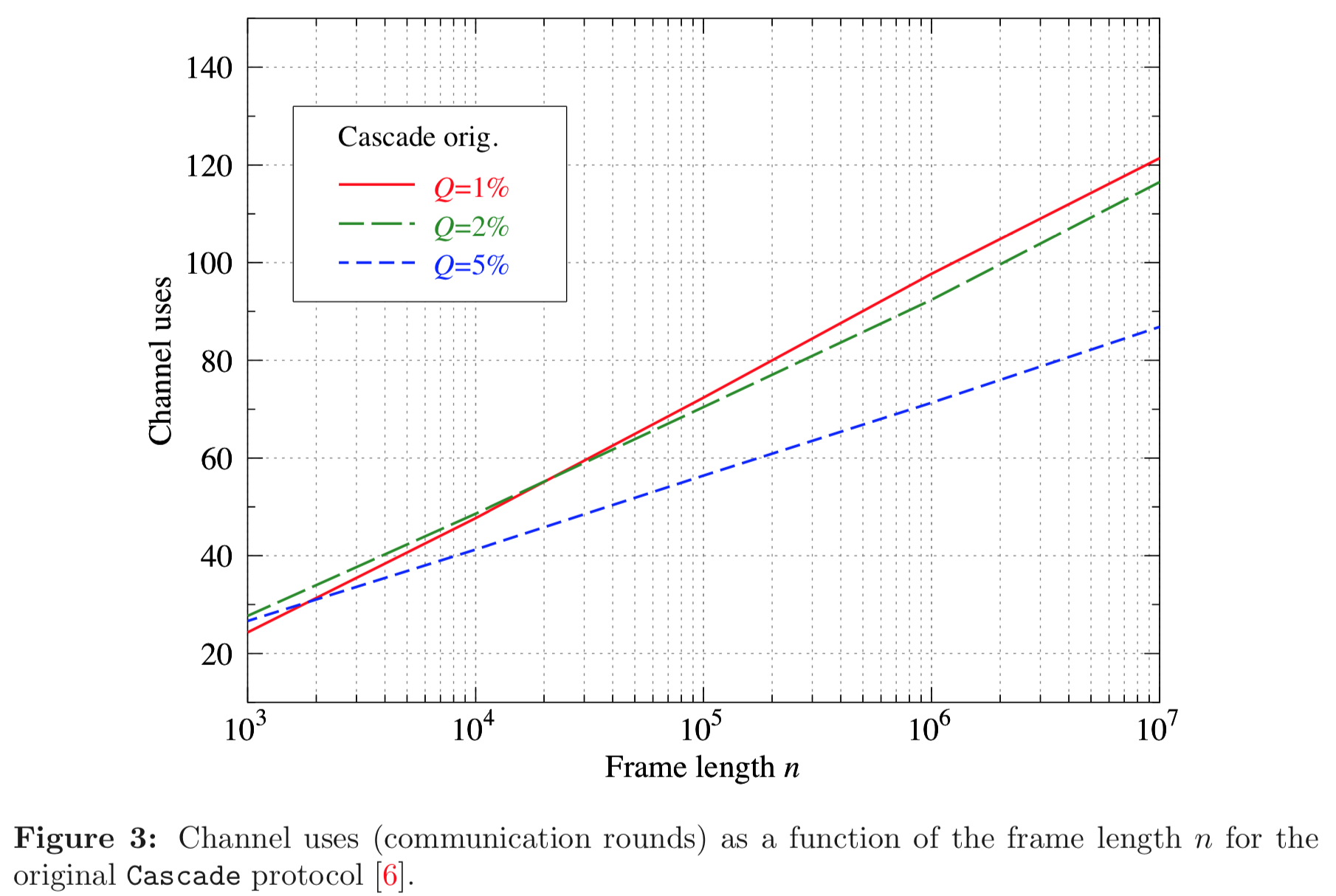
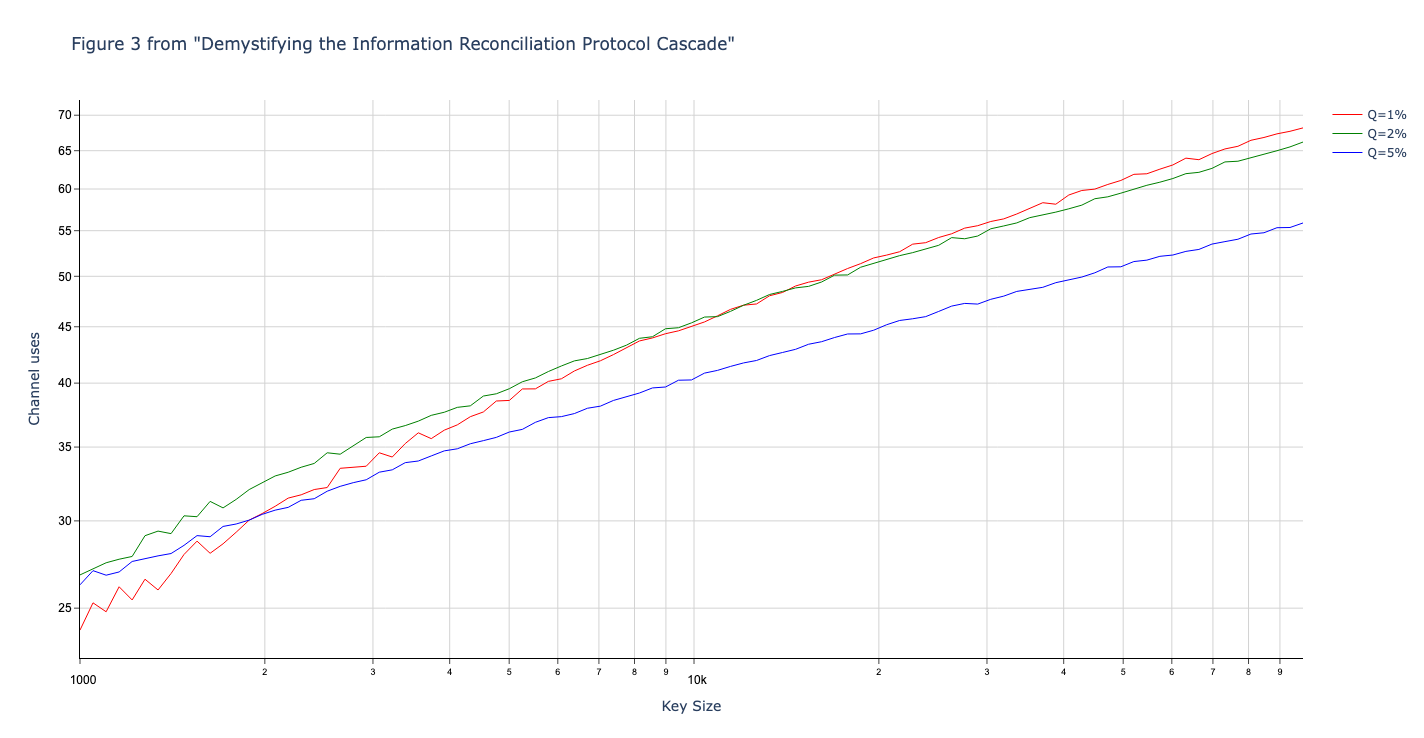
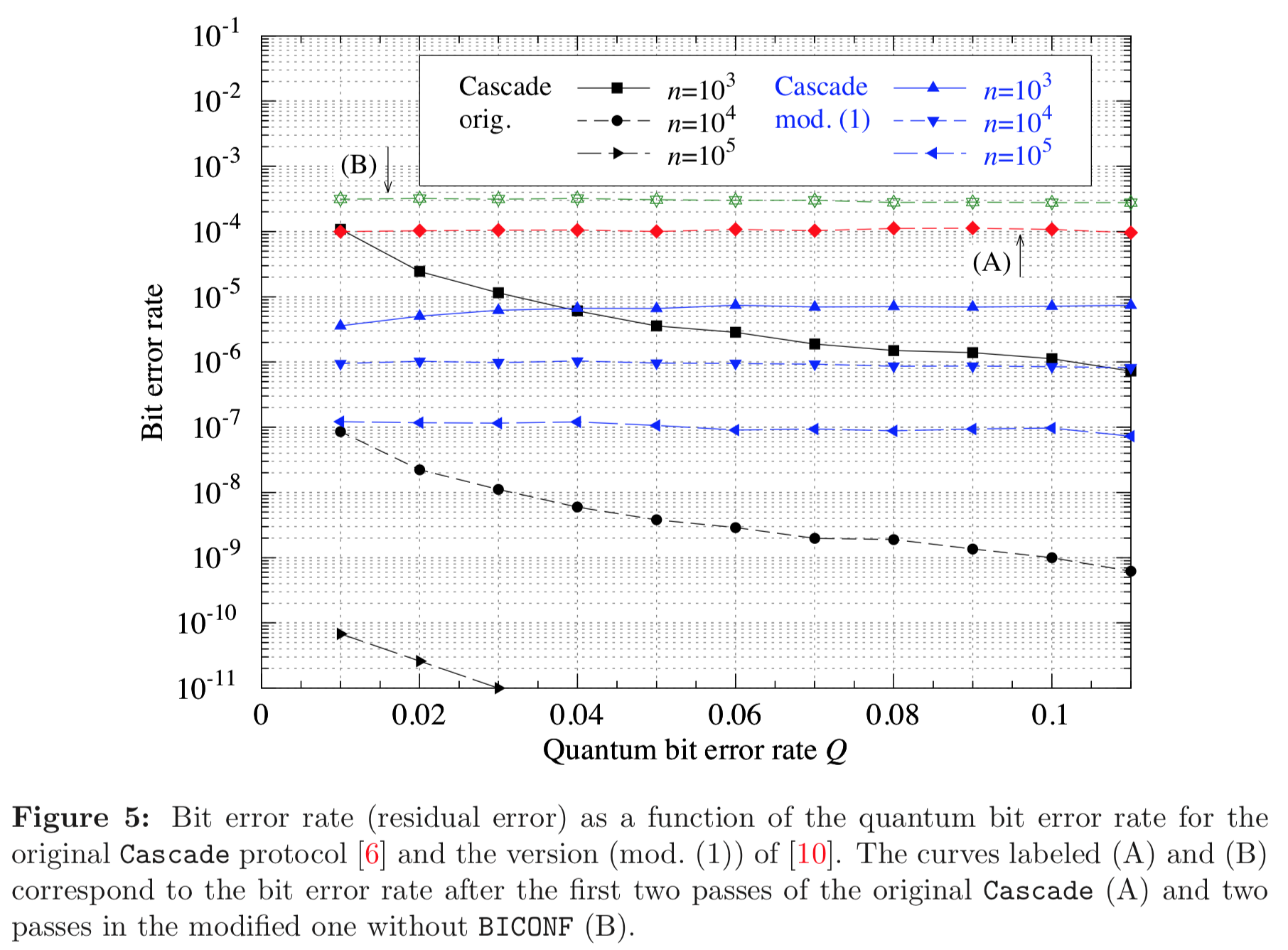
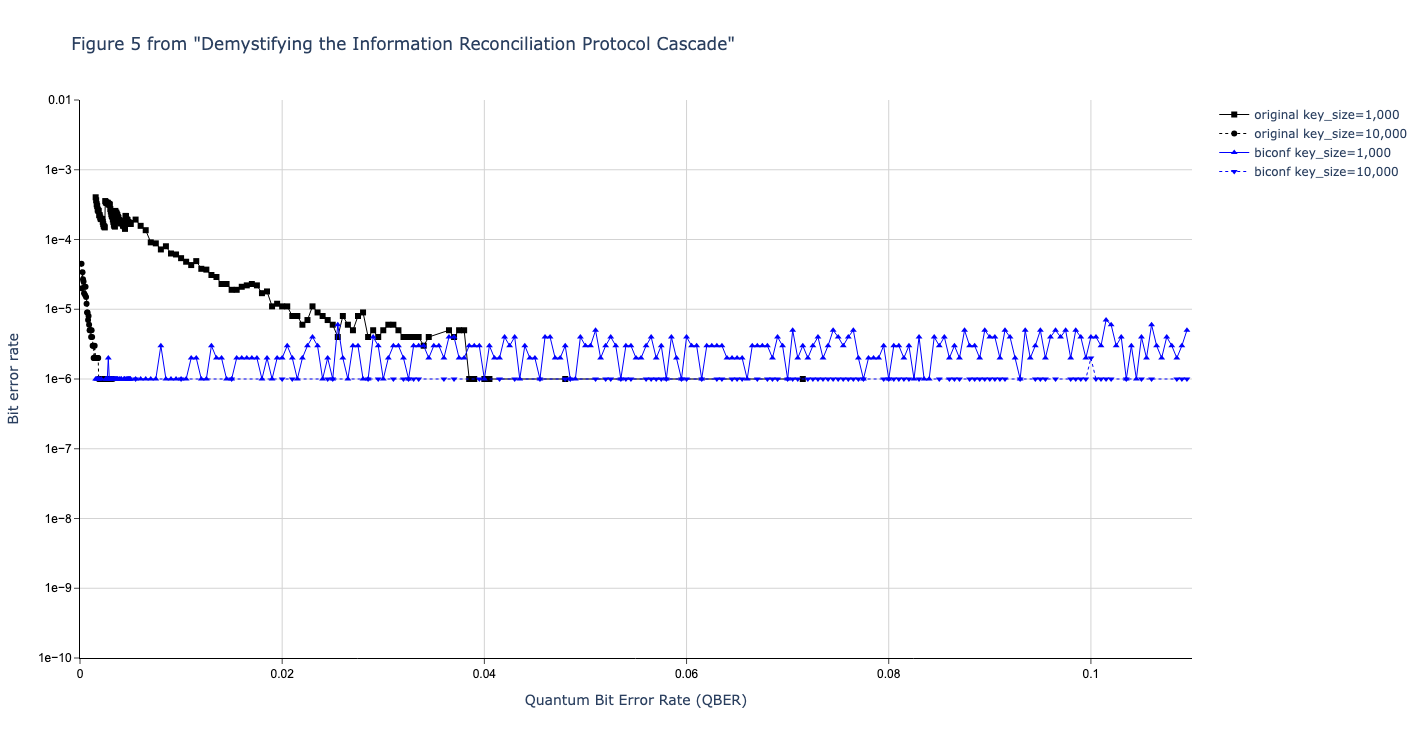
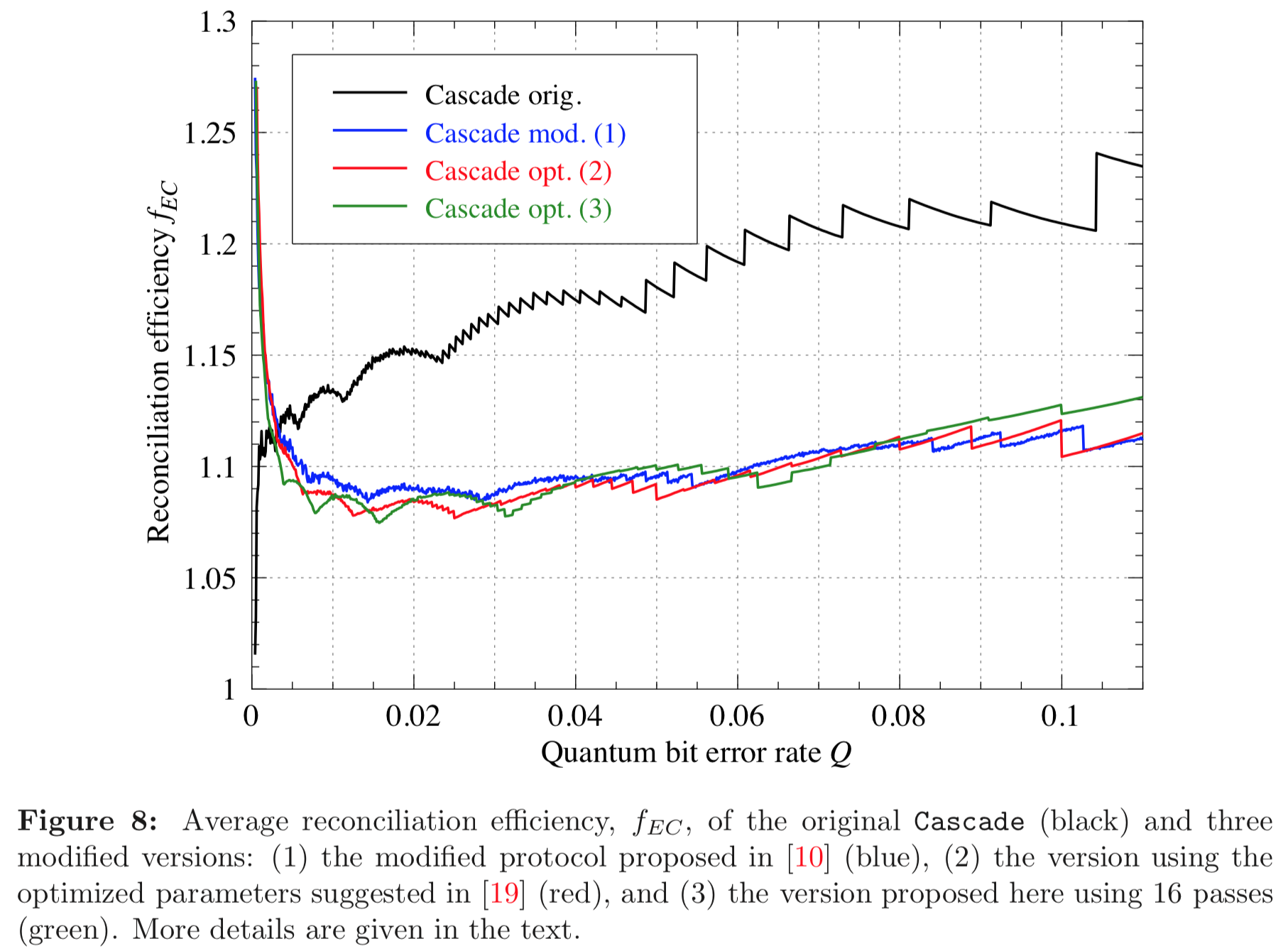
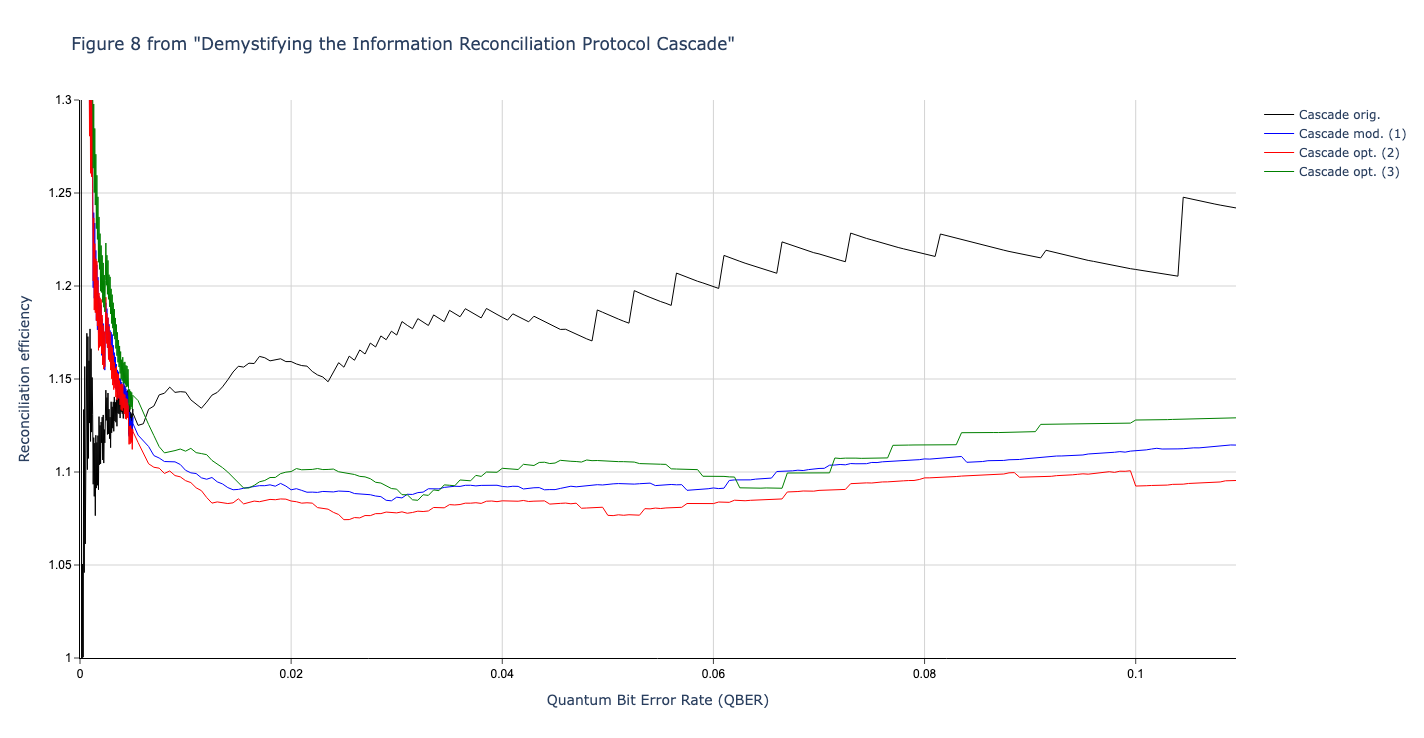
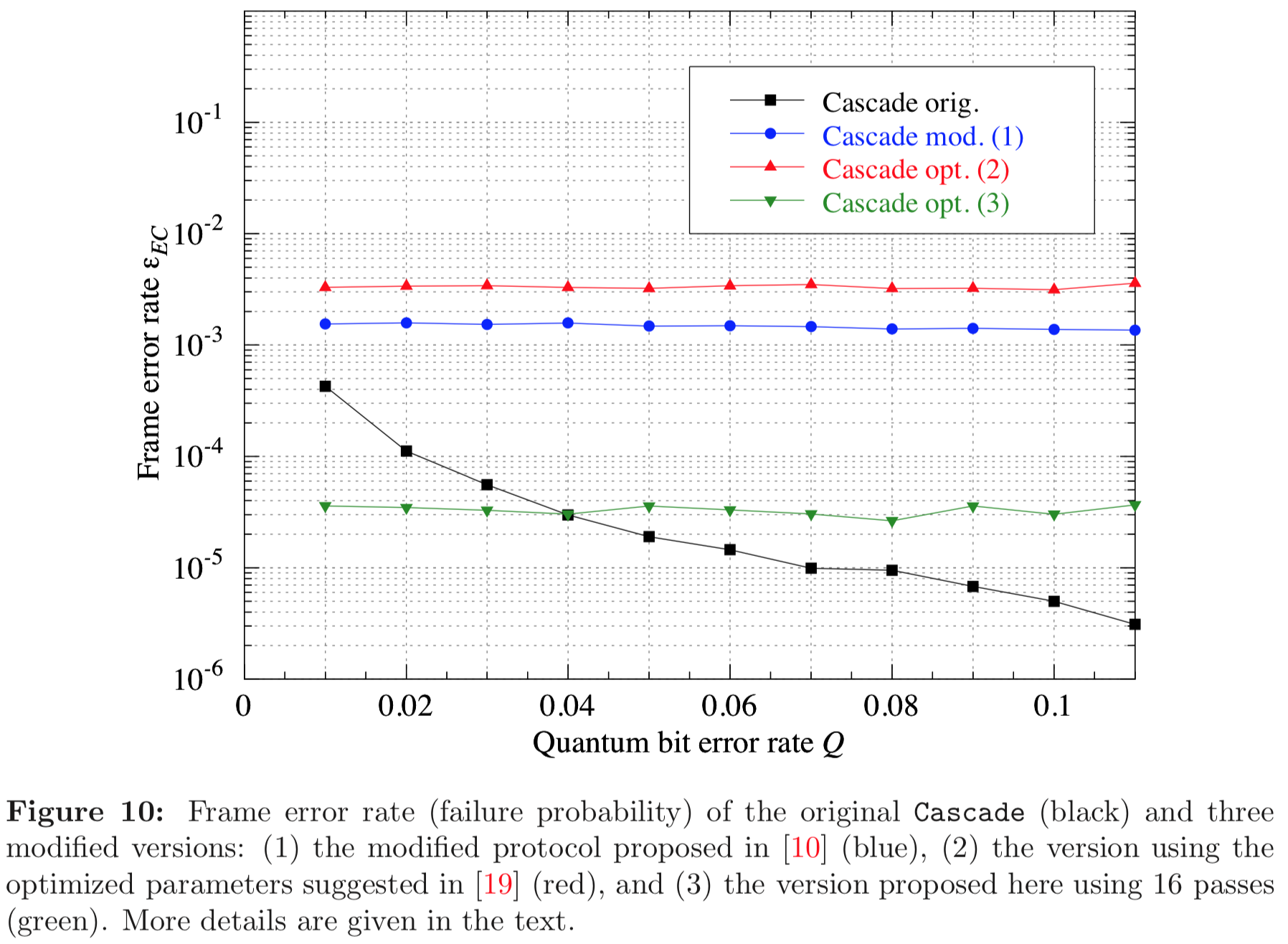
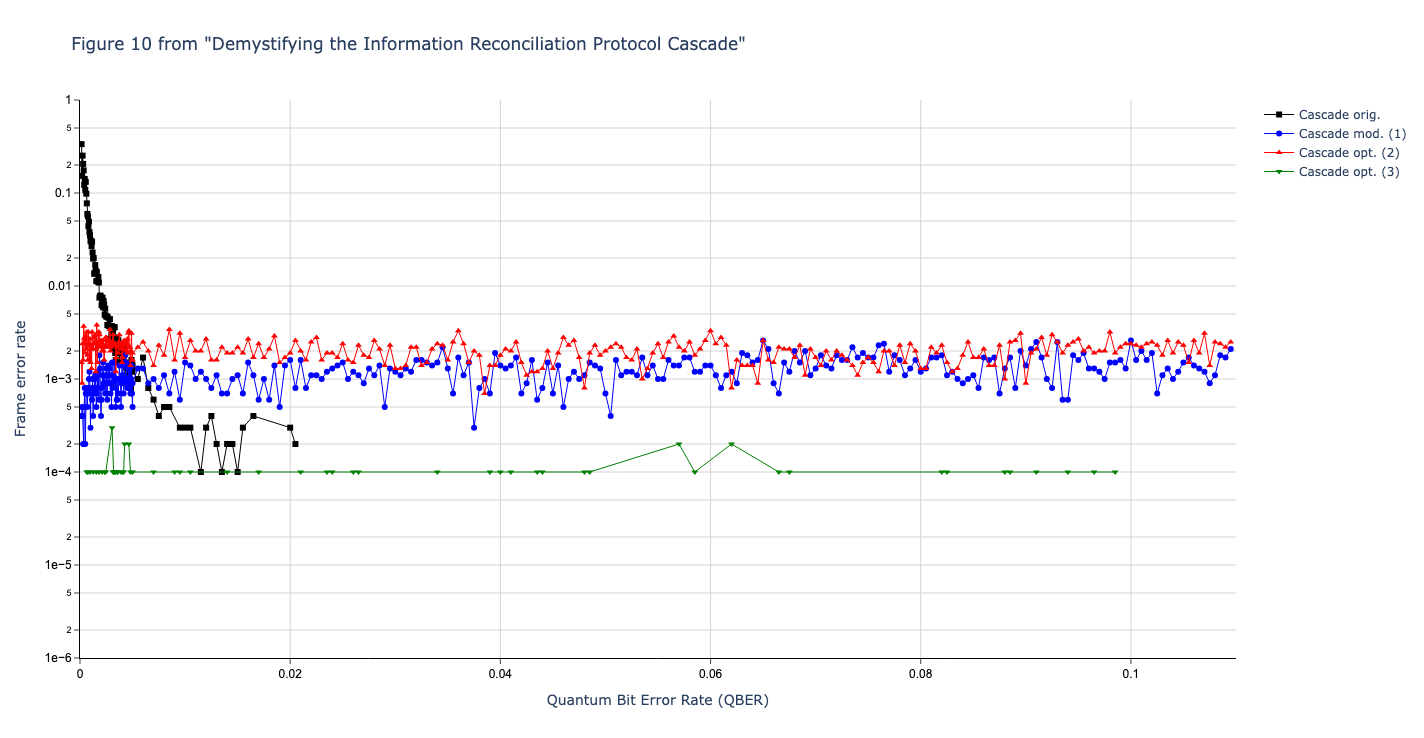
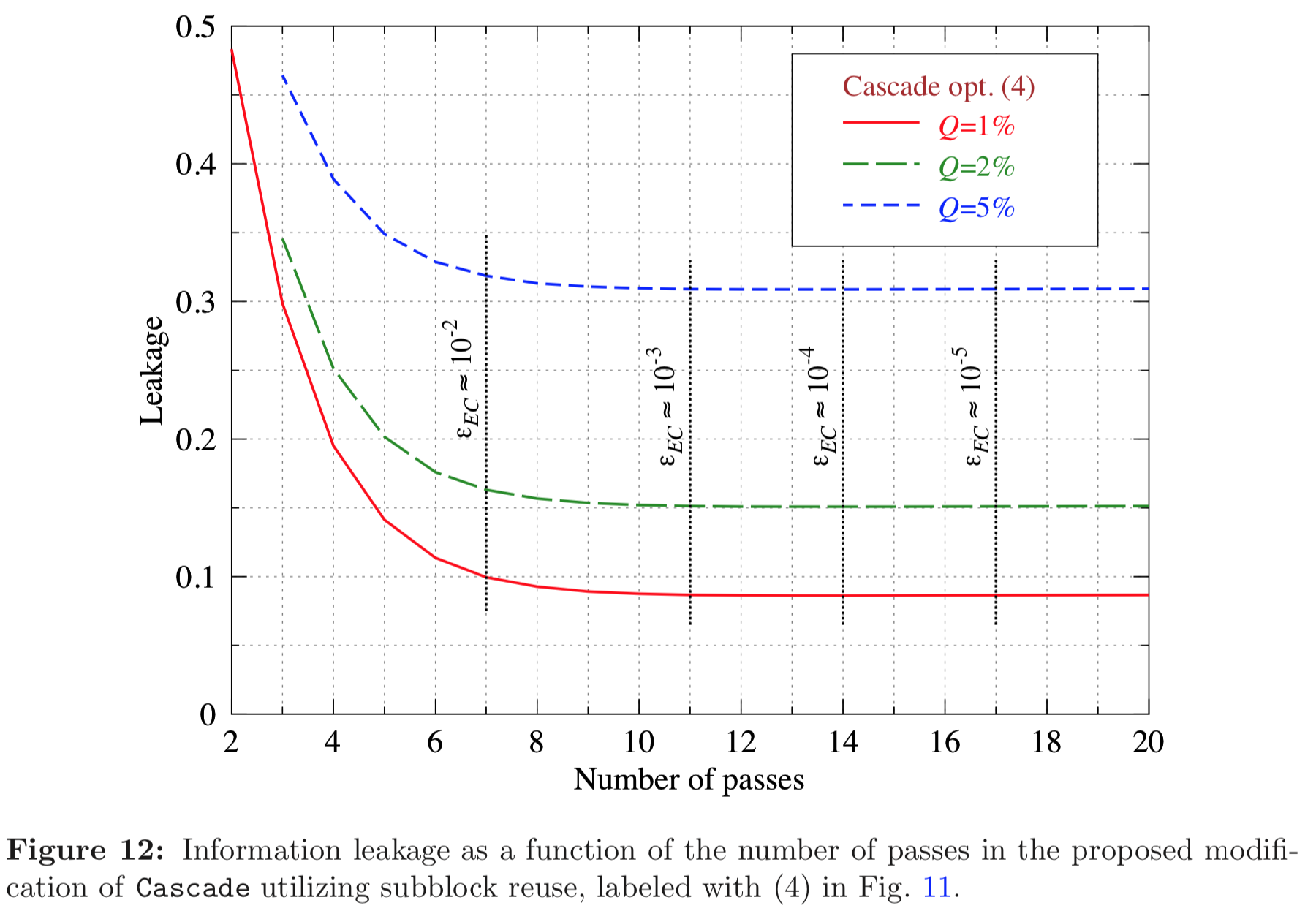
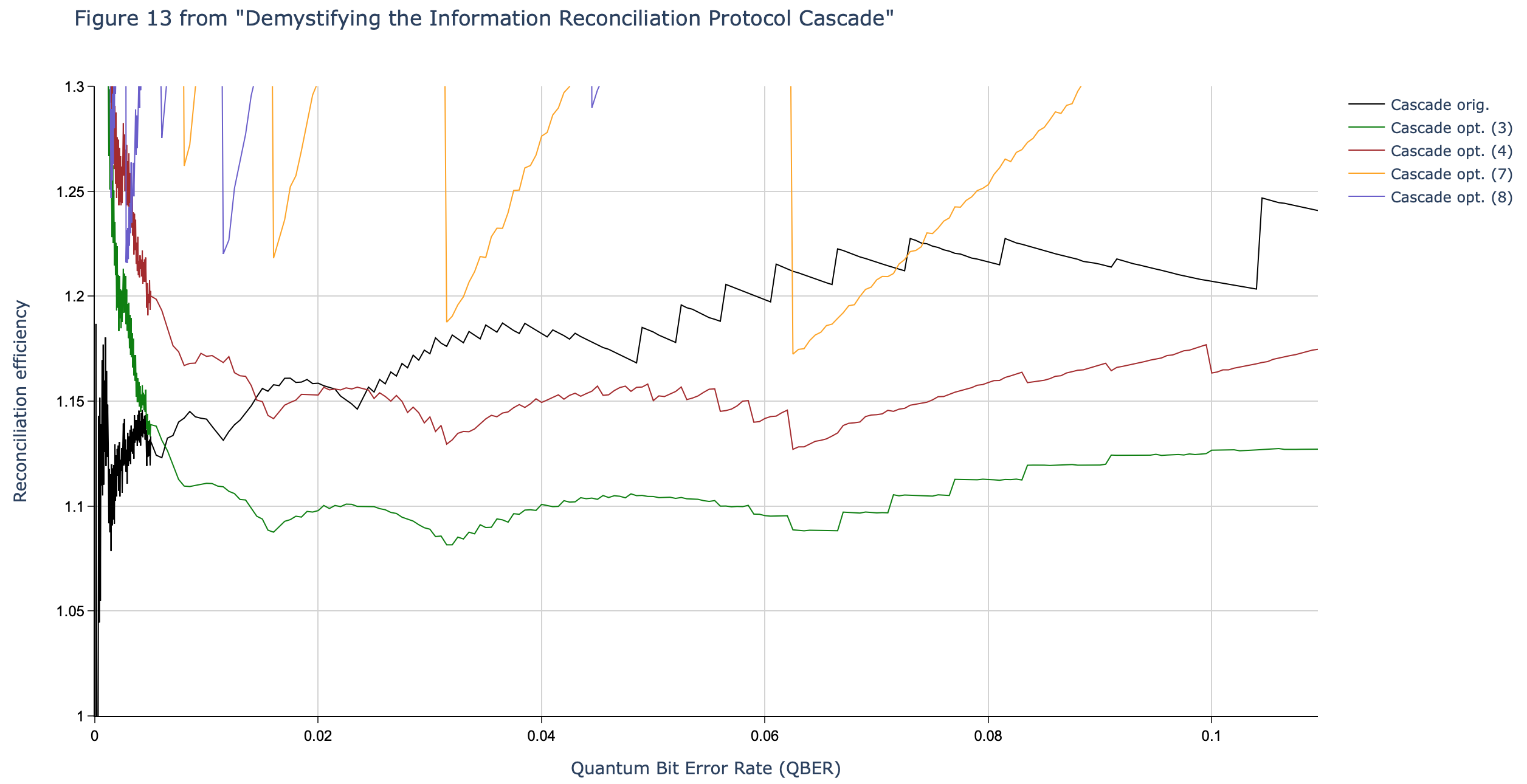
Results reported in paper:
Results produced by Cascade-CPP:
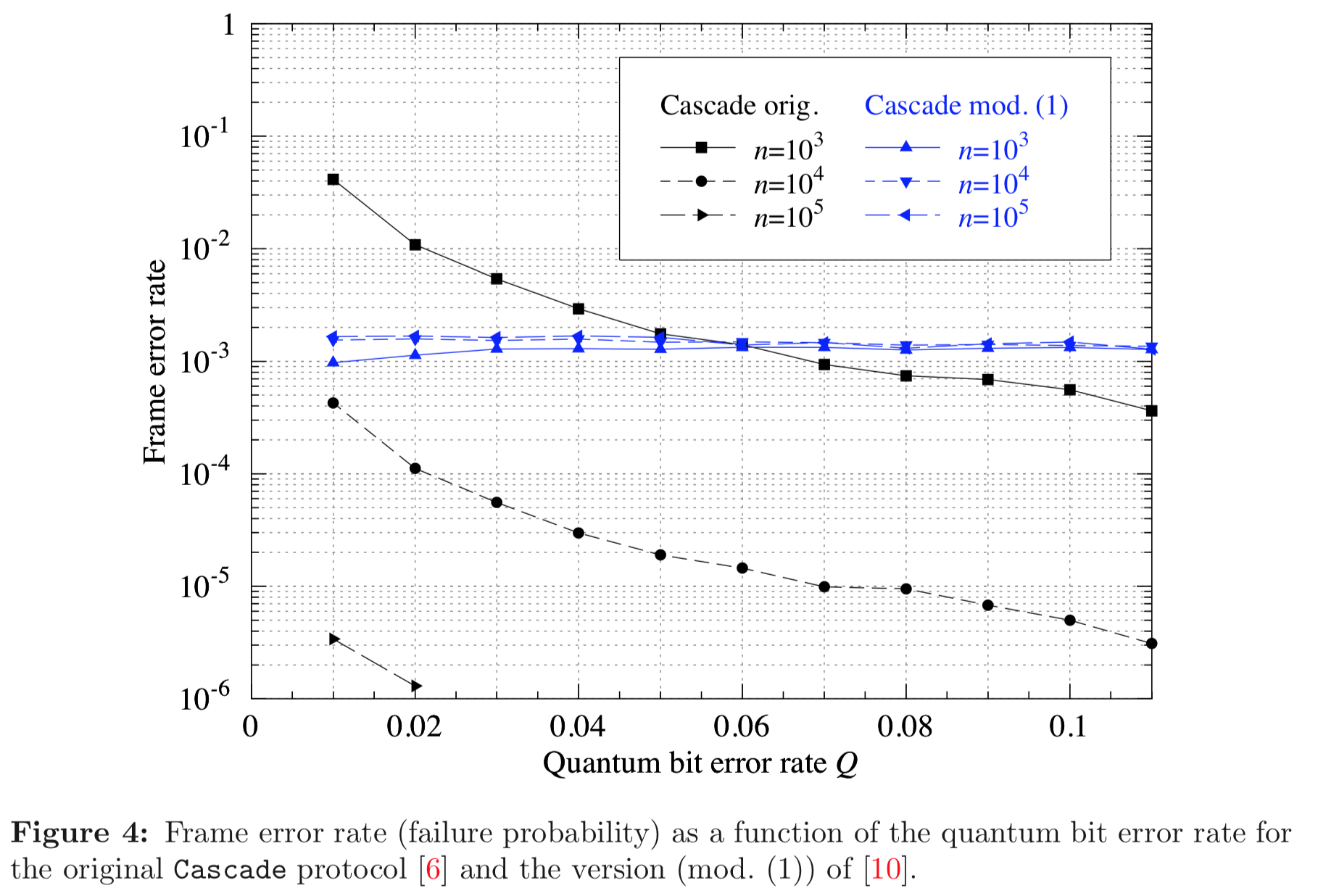
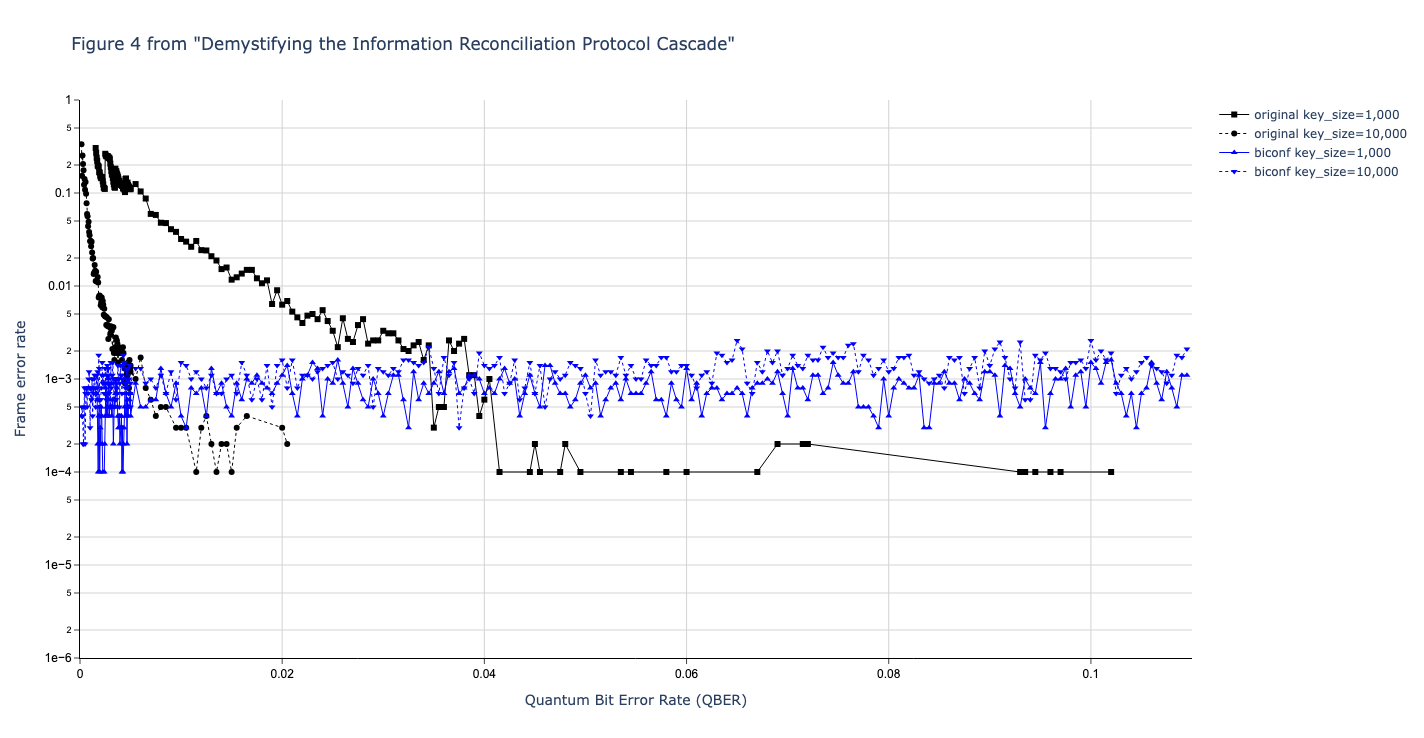
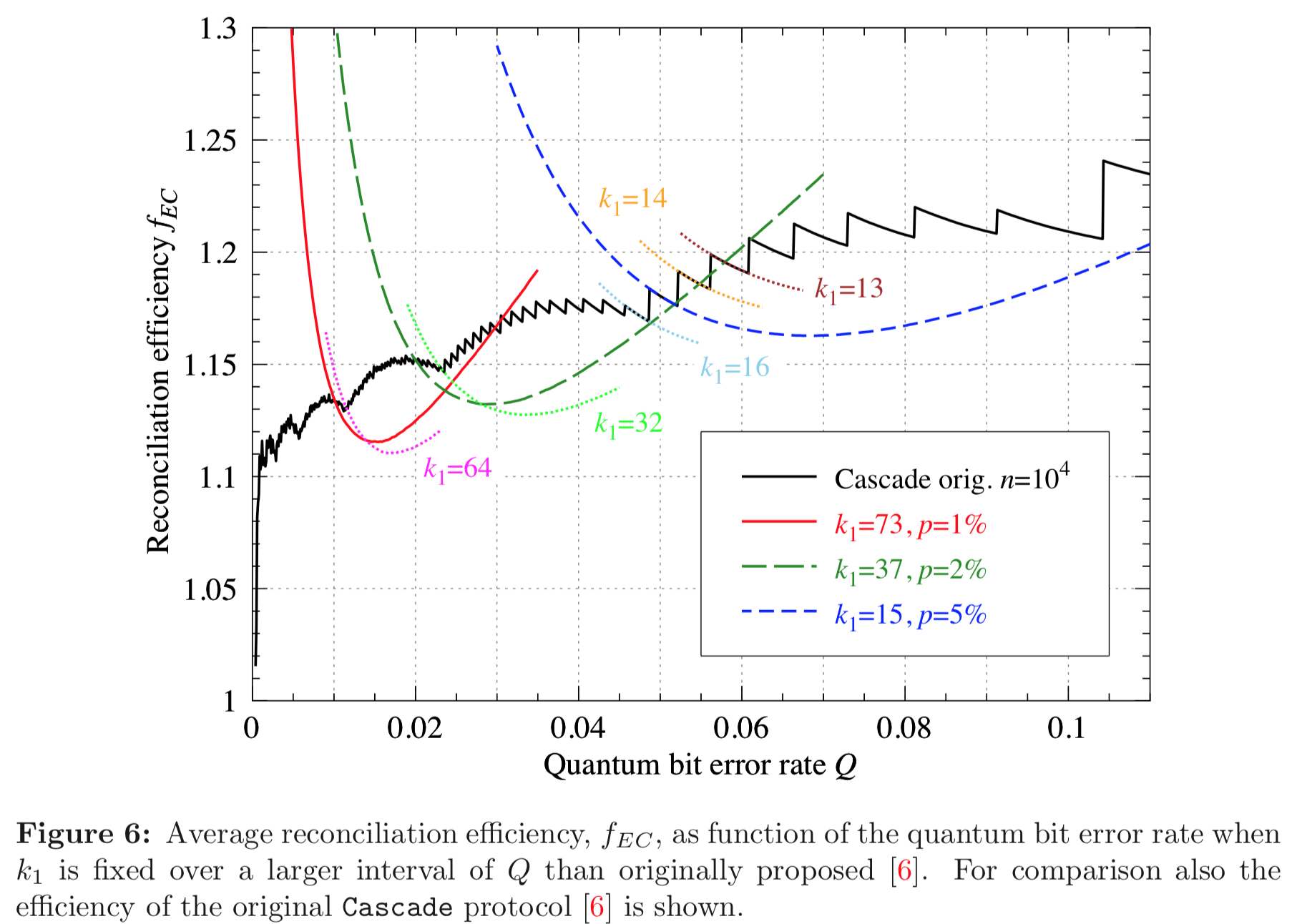
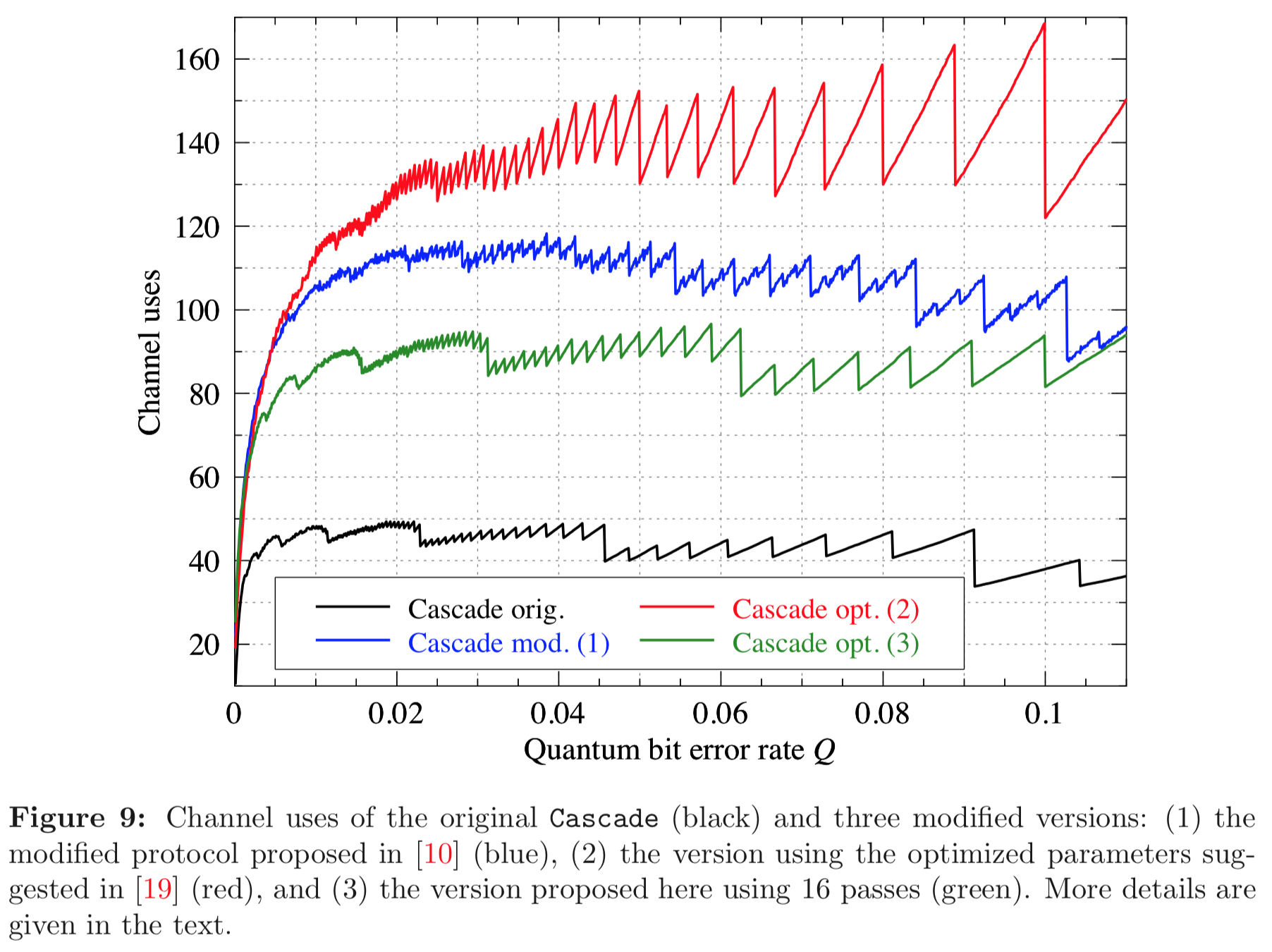
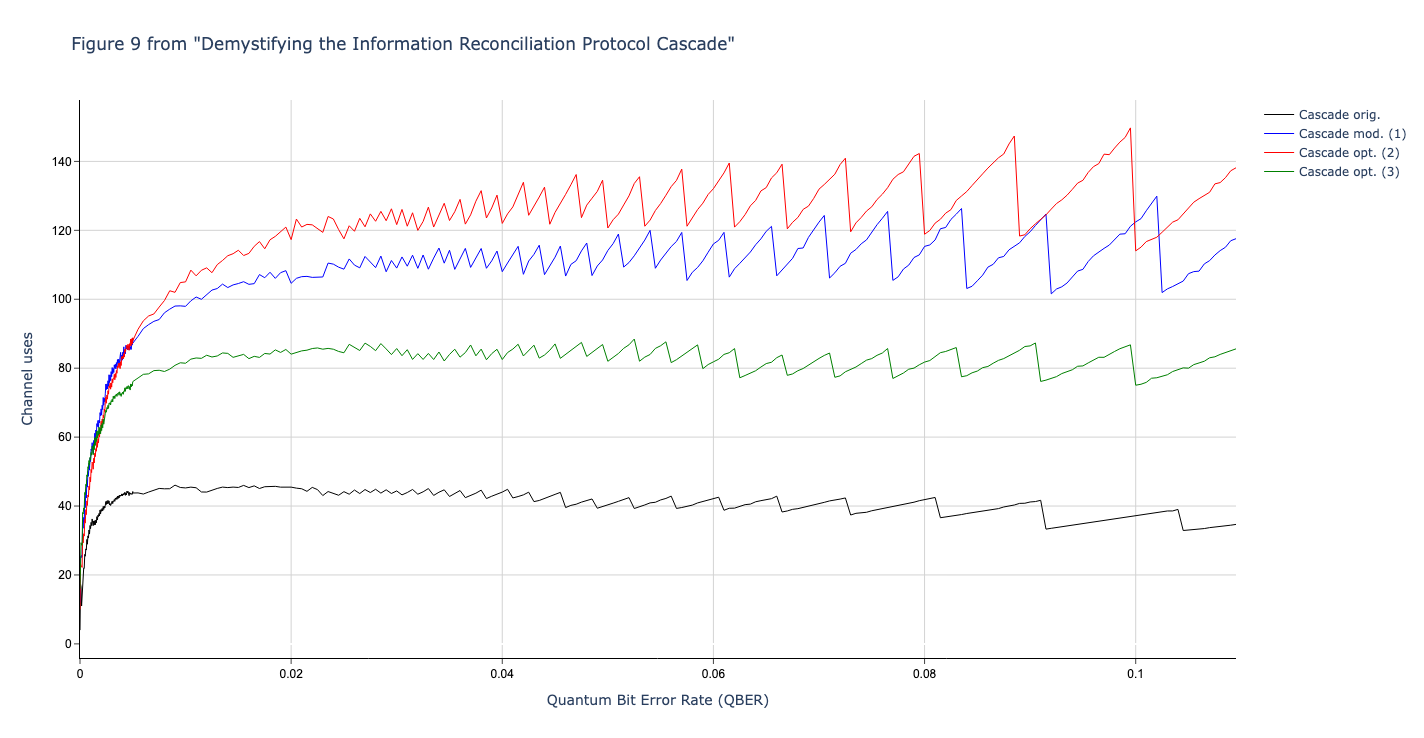
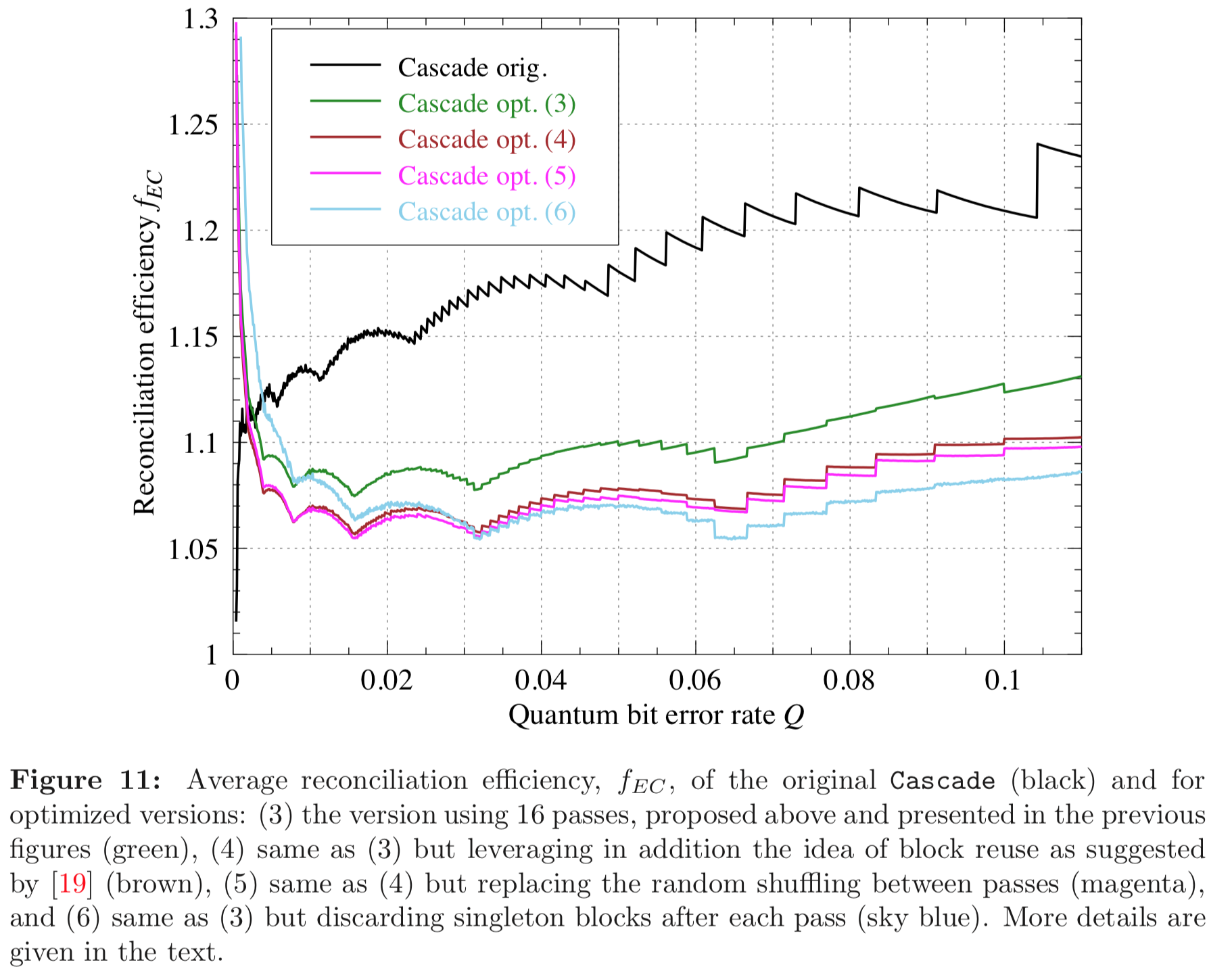
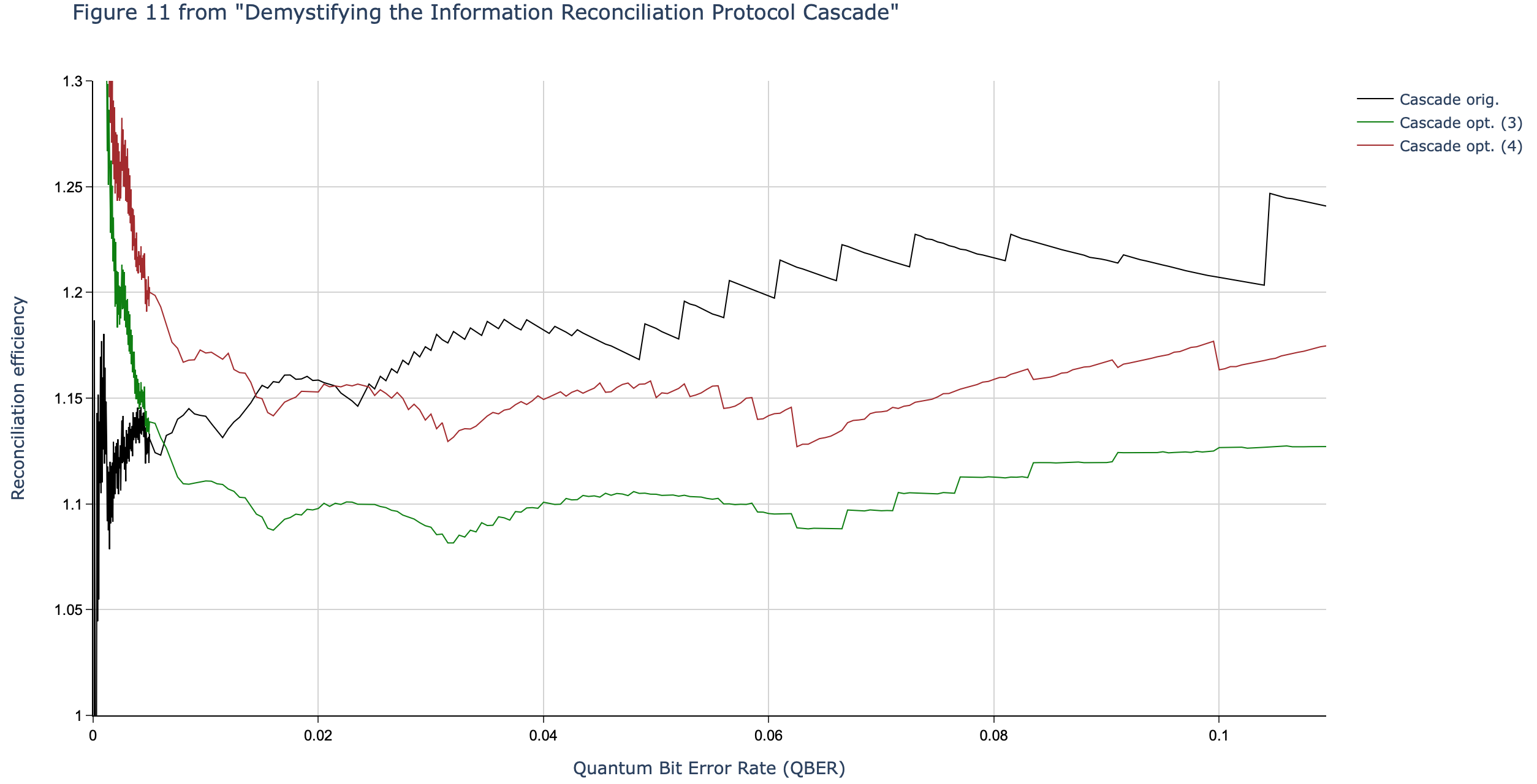
Results reported in paper:
Results produced by Cascade-CPP:
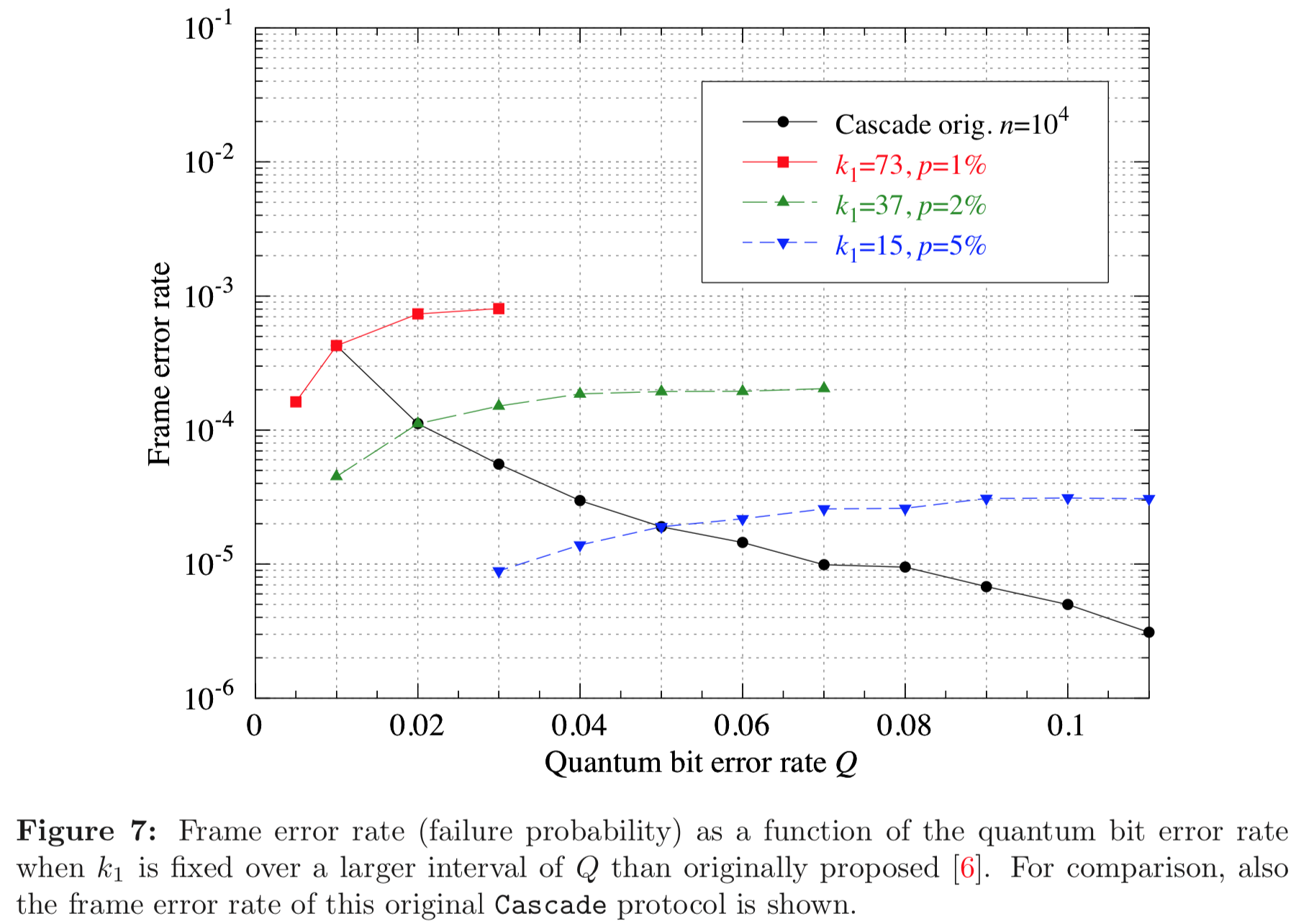
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results not produced by Cascade-CPP.
Results reported in paper:
Results not produced by Cascade-CPP.
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results produced by Cascade-CPP:
Results reported in paper:
Results not produced by Cascade-CPP.
Results reported in paper:
Results produced by Cascade-CPP:
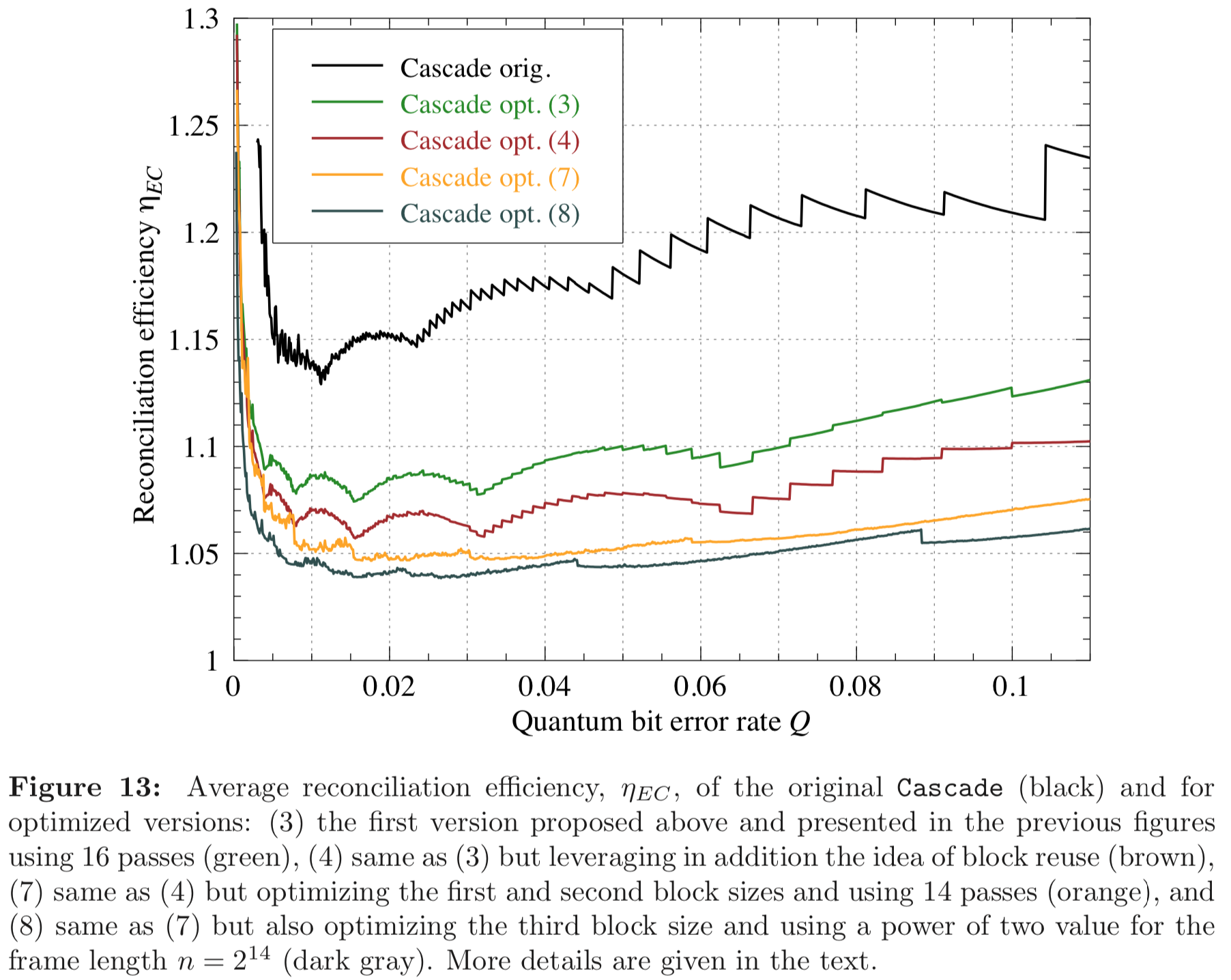
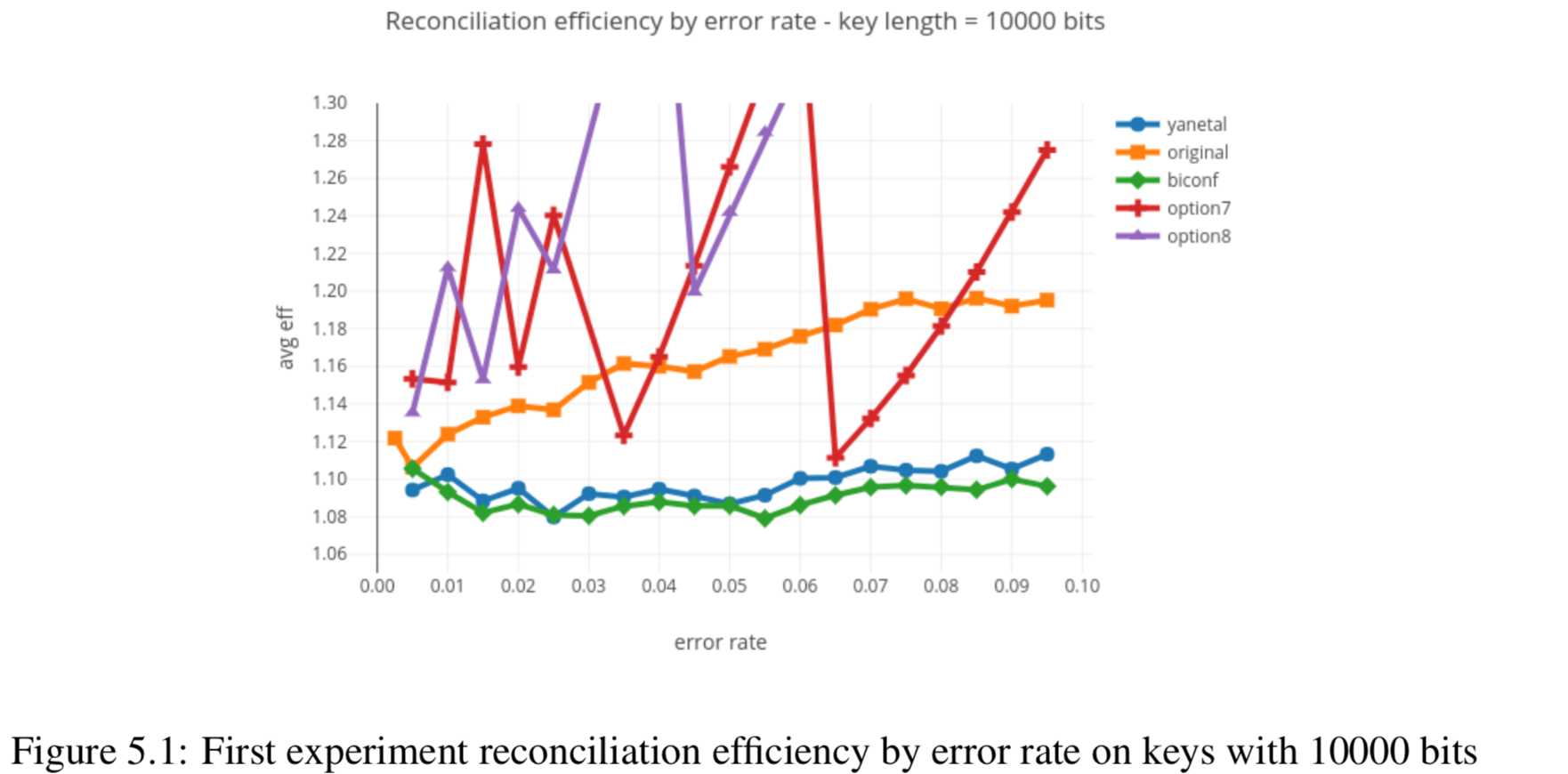
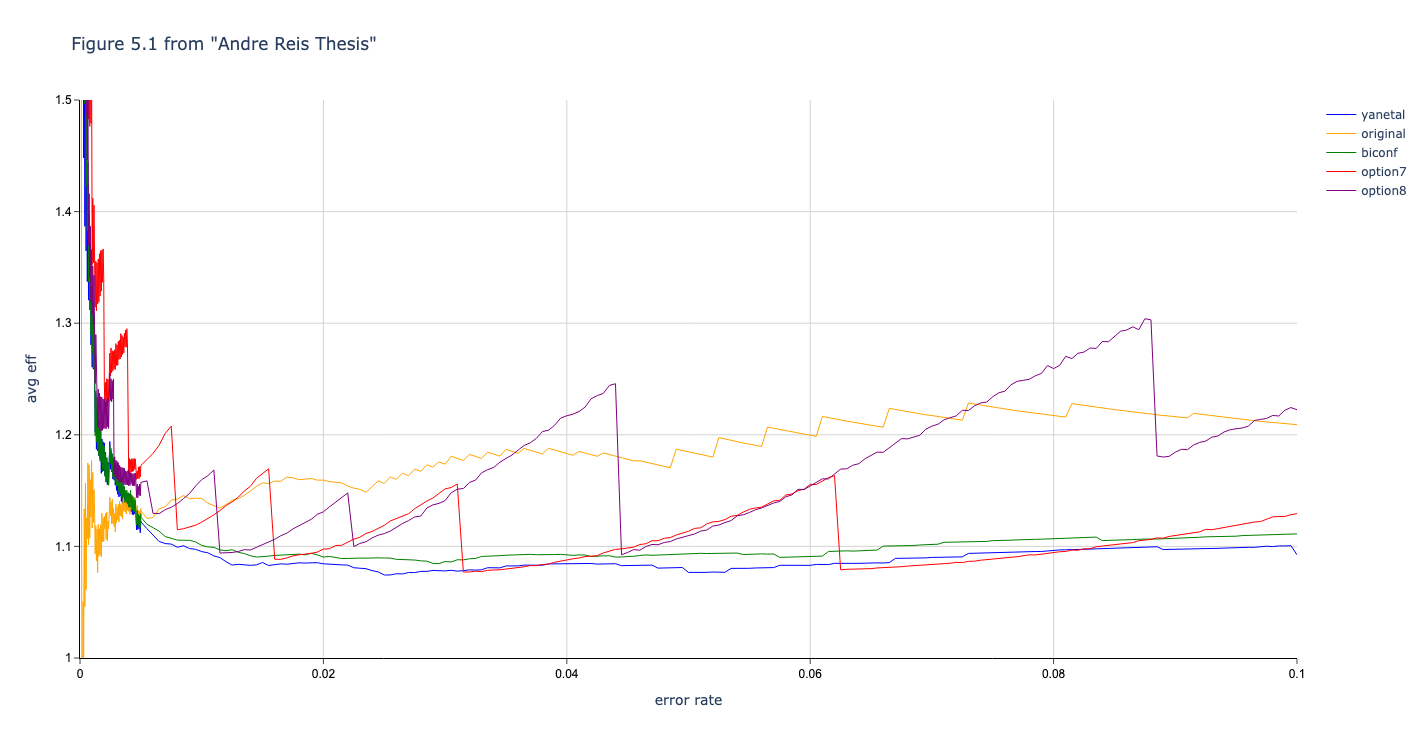
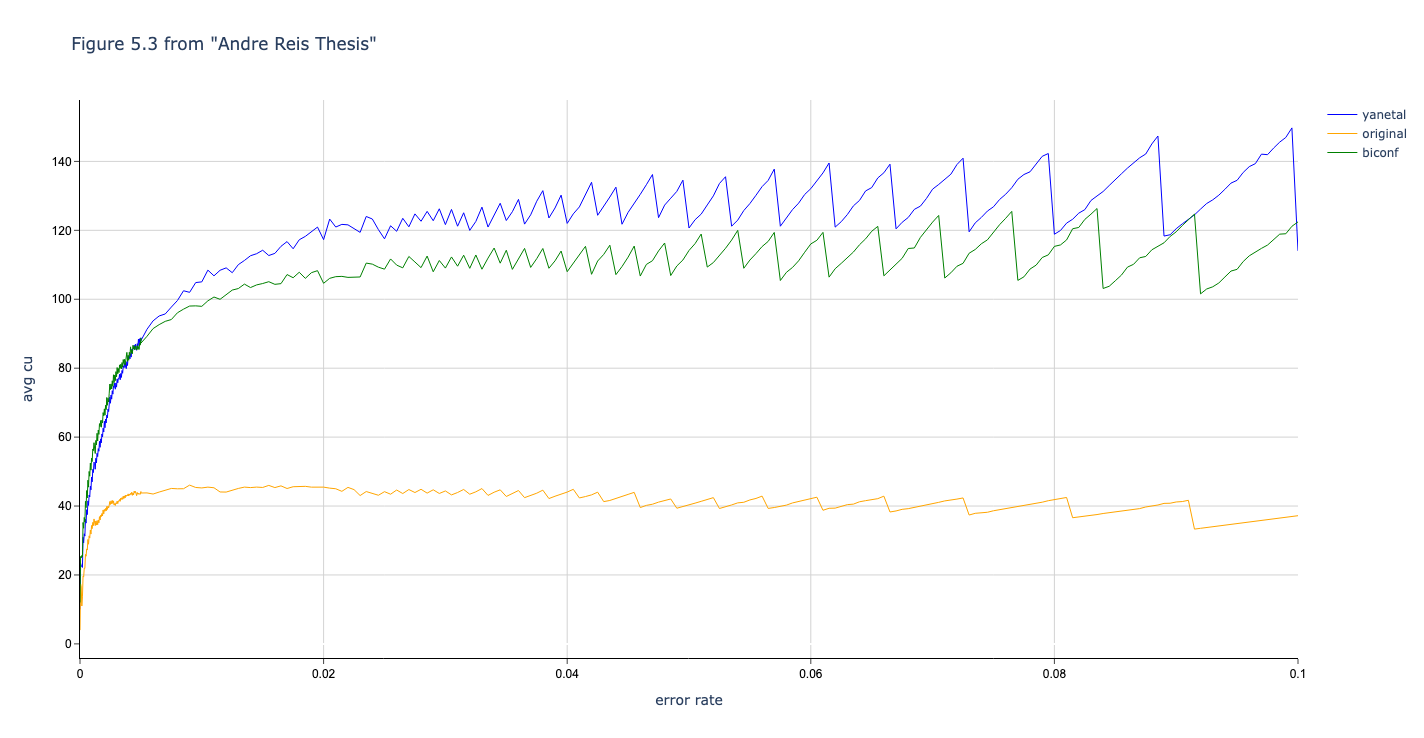
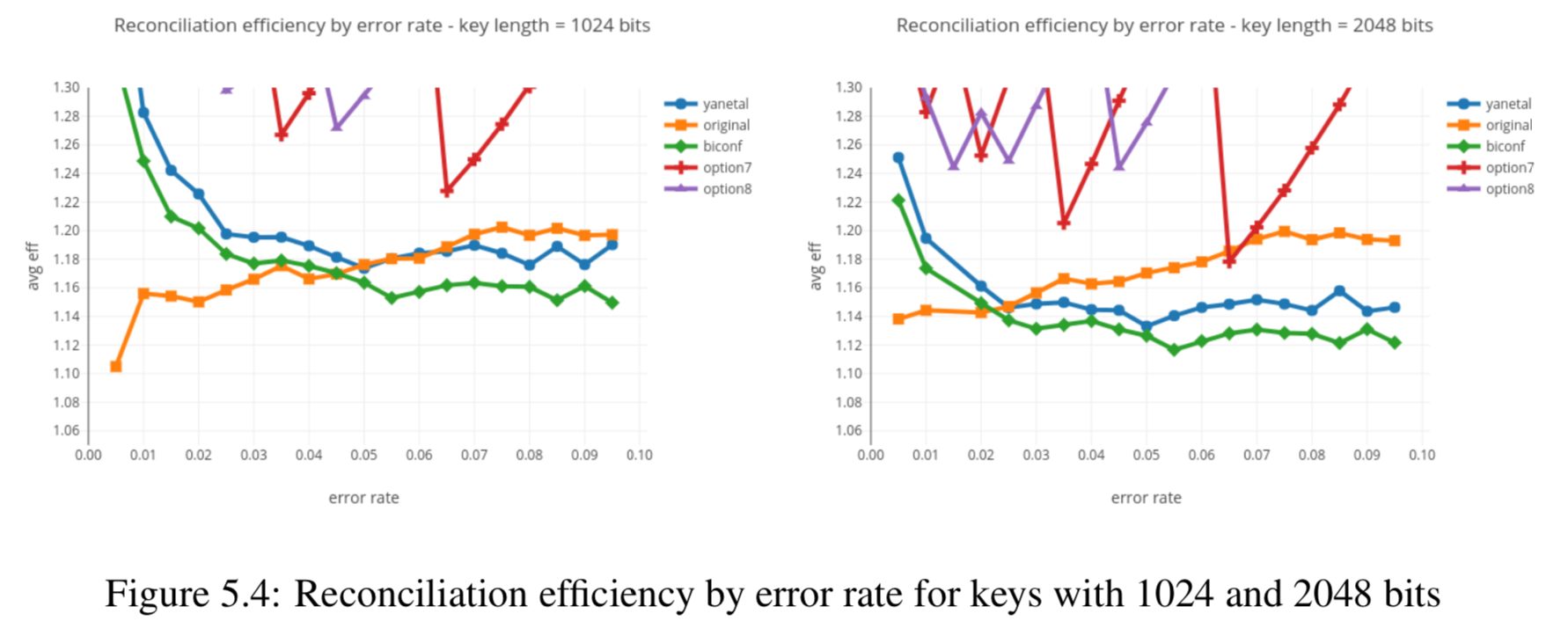
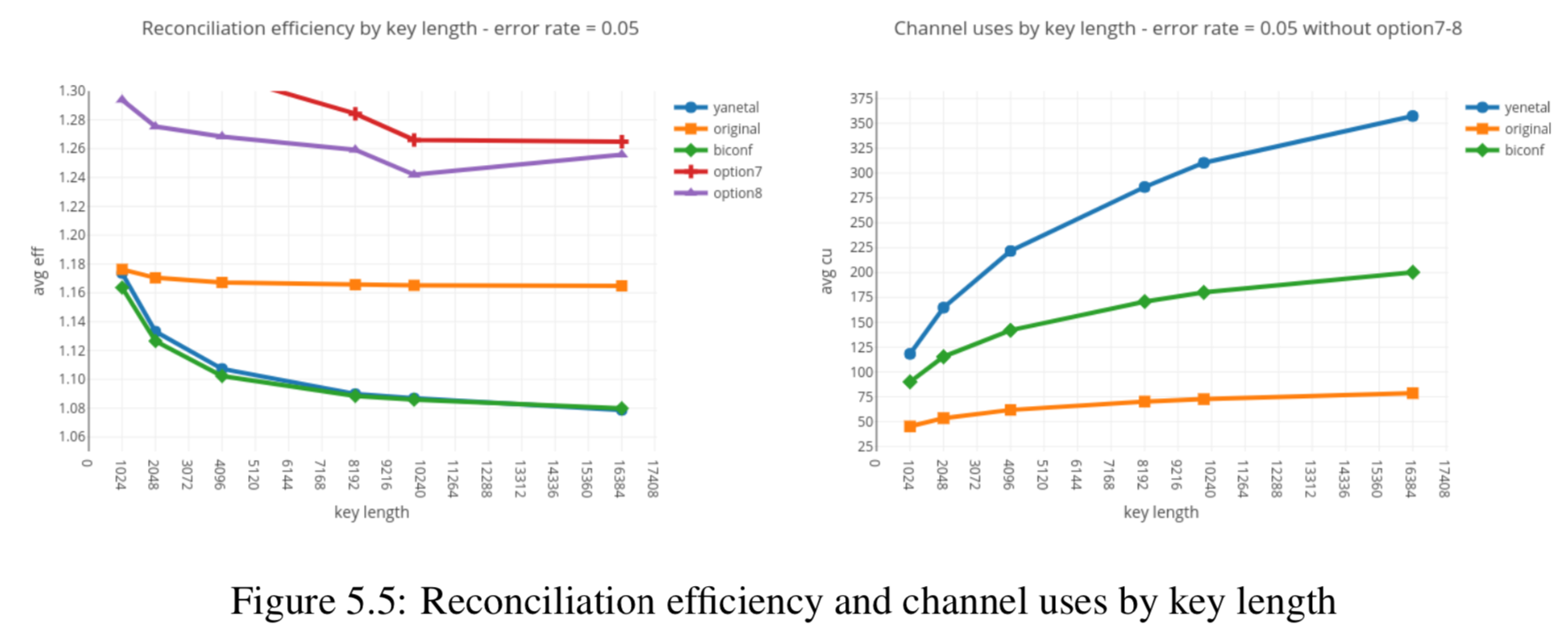
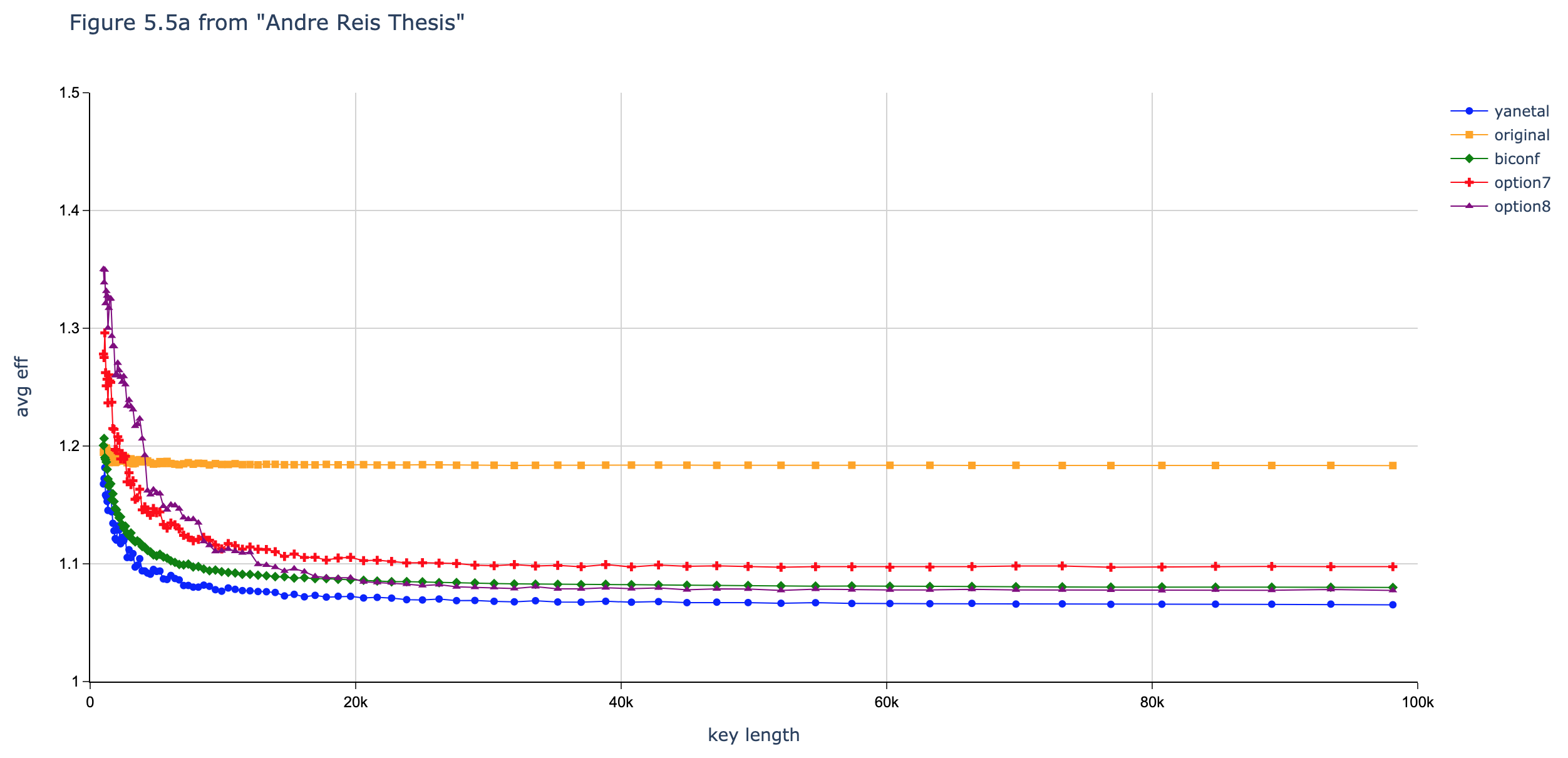
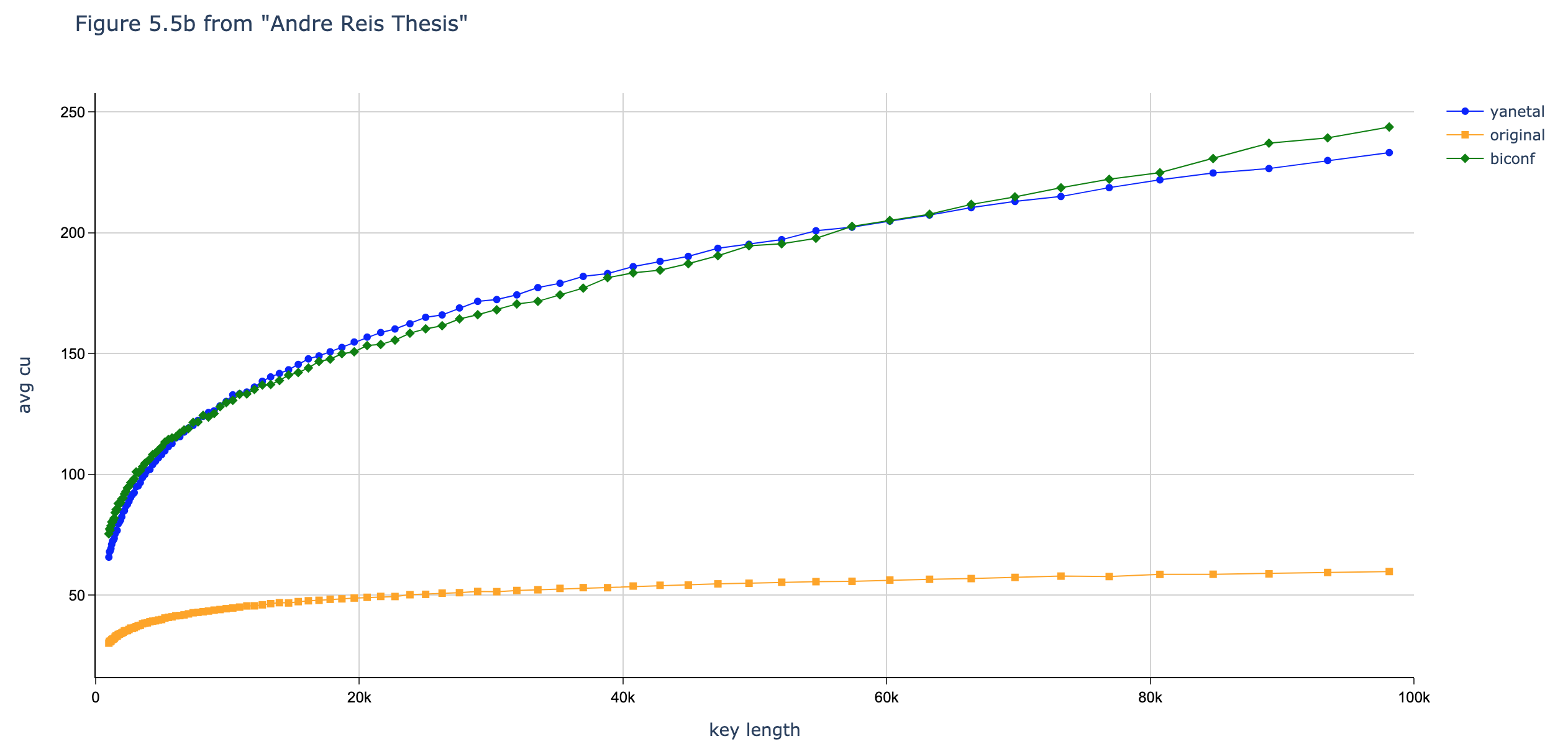
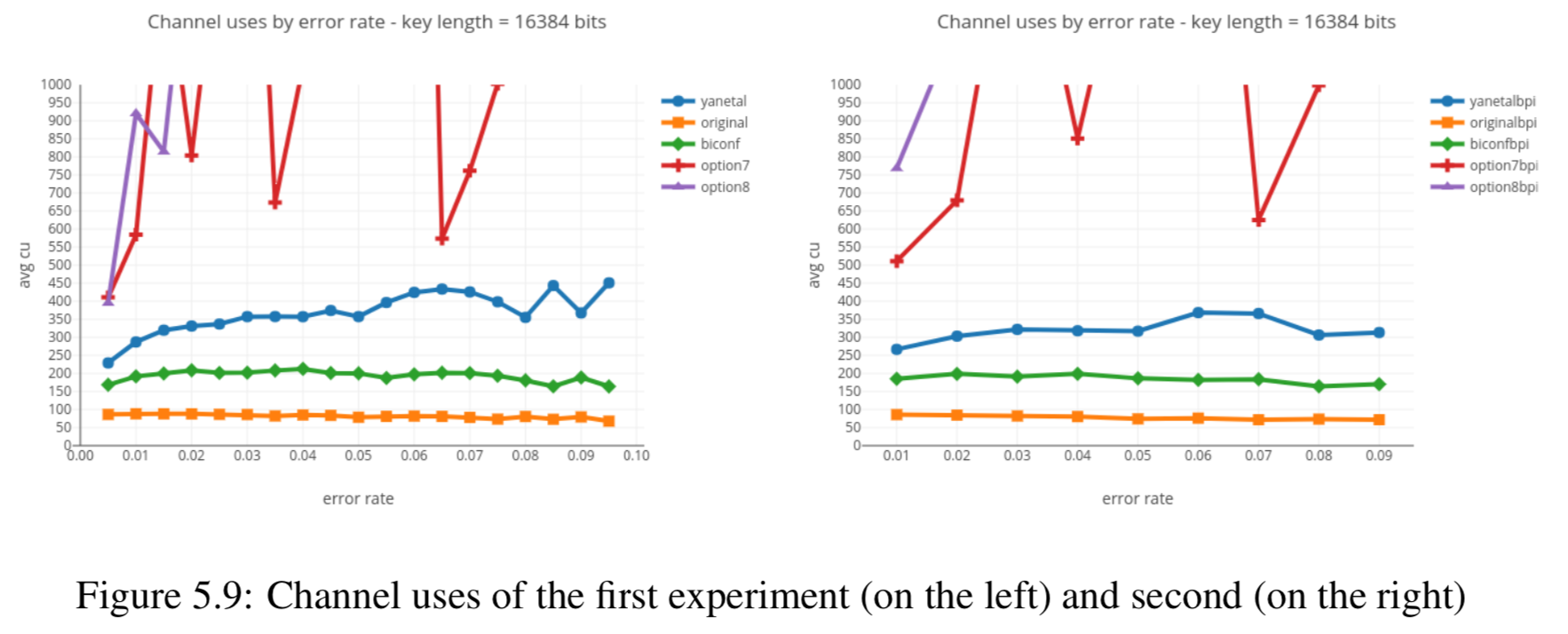
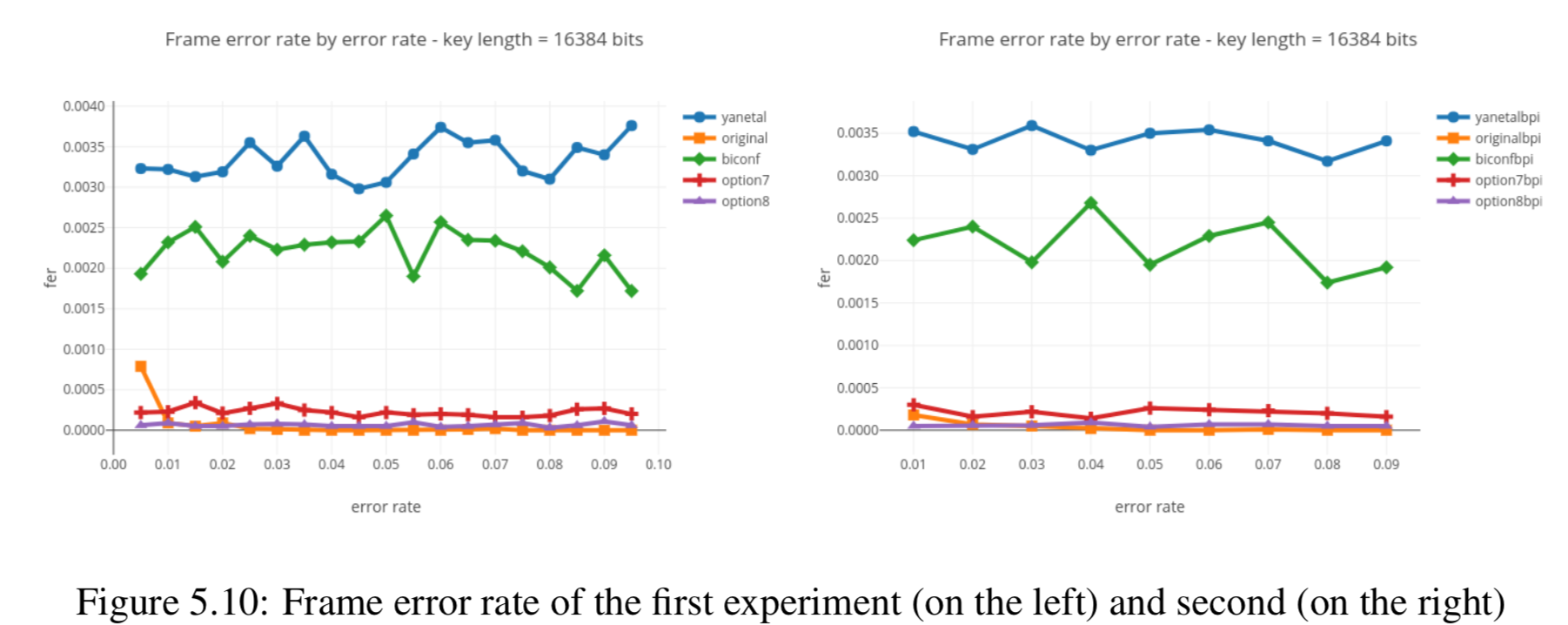
In this section we compare the data produced by Cascade-CPP with the results reported in:
[Quantum Key Distribution Post Processing - A study on the Information Reconciliation Cascade Protocol.](https://repositorio-aberto.up.pt/bitstream/10216/121965/2/347567.pdf.
André Reis.
Master’s Thesis, Faculdade de Engenharia da Universidade do Porto, Jul 2019.
Results reported in thesis:
Results produced by Cascade-CPP:
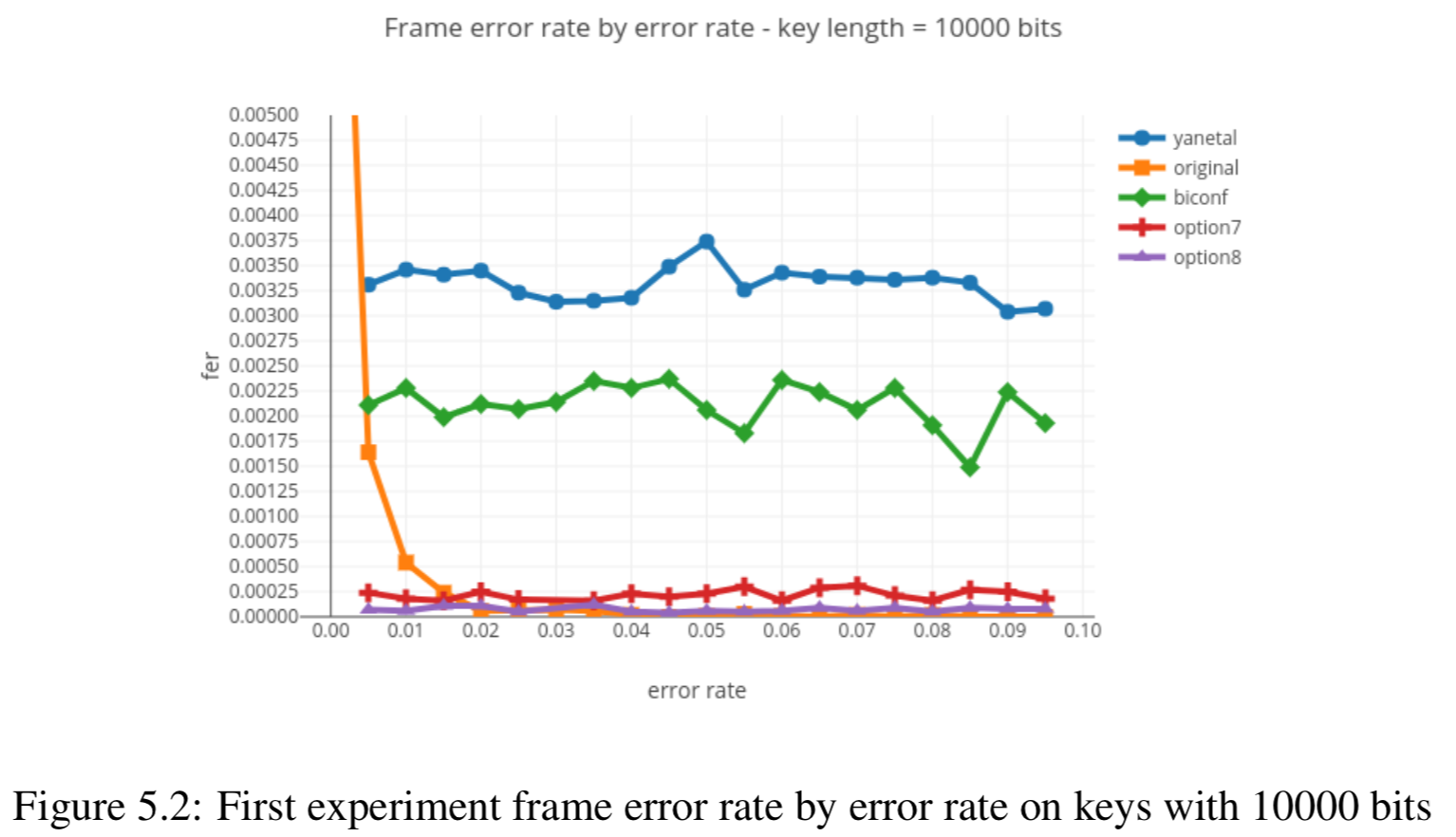
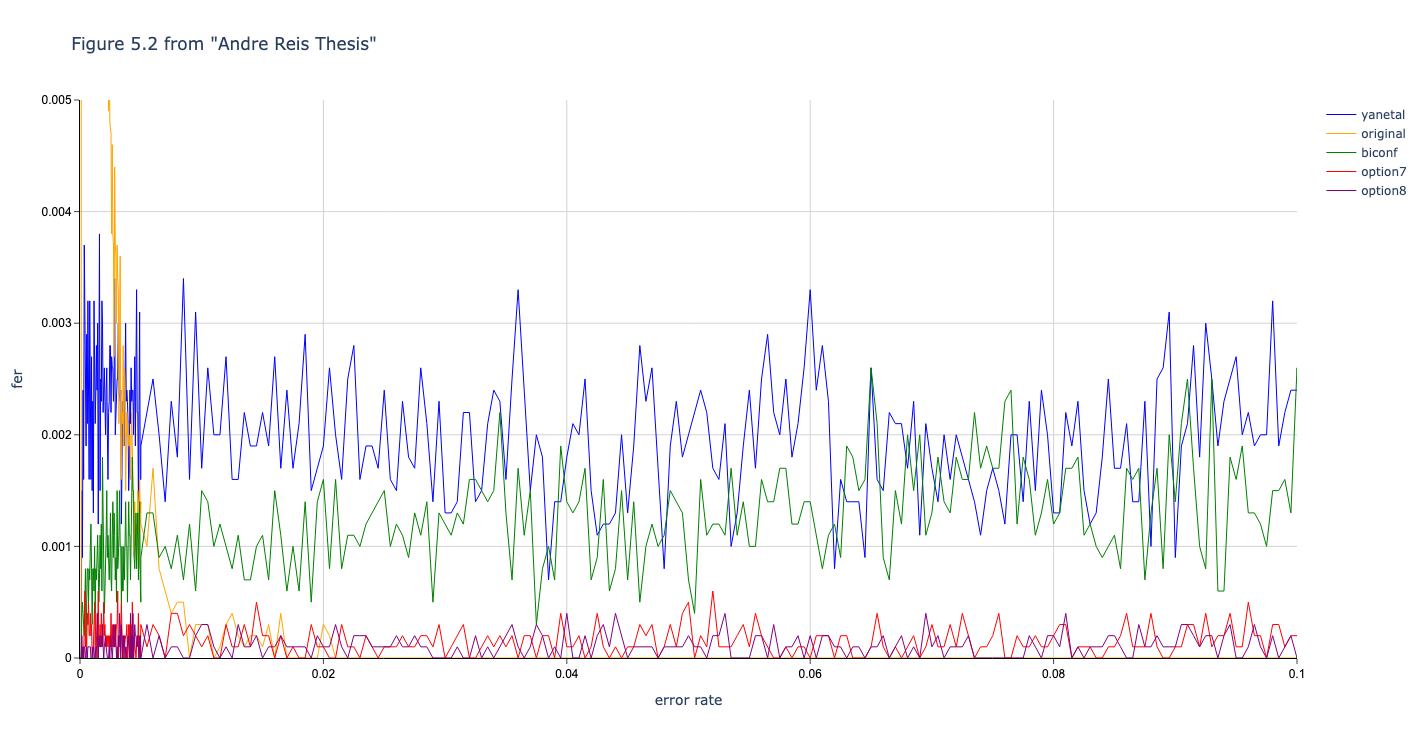
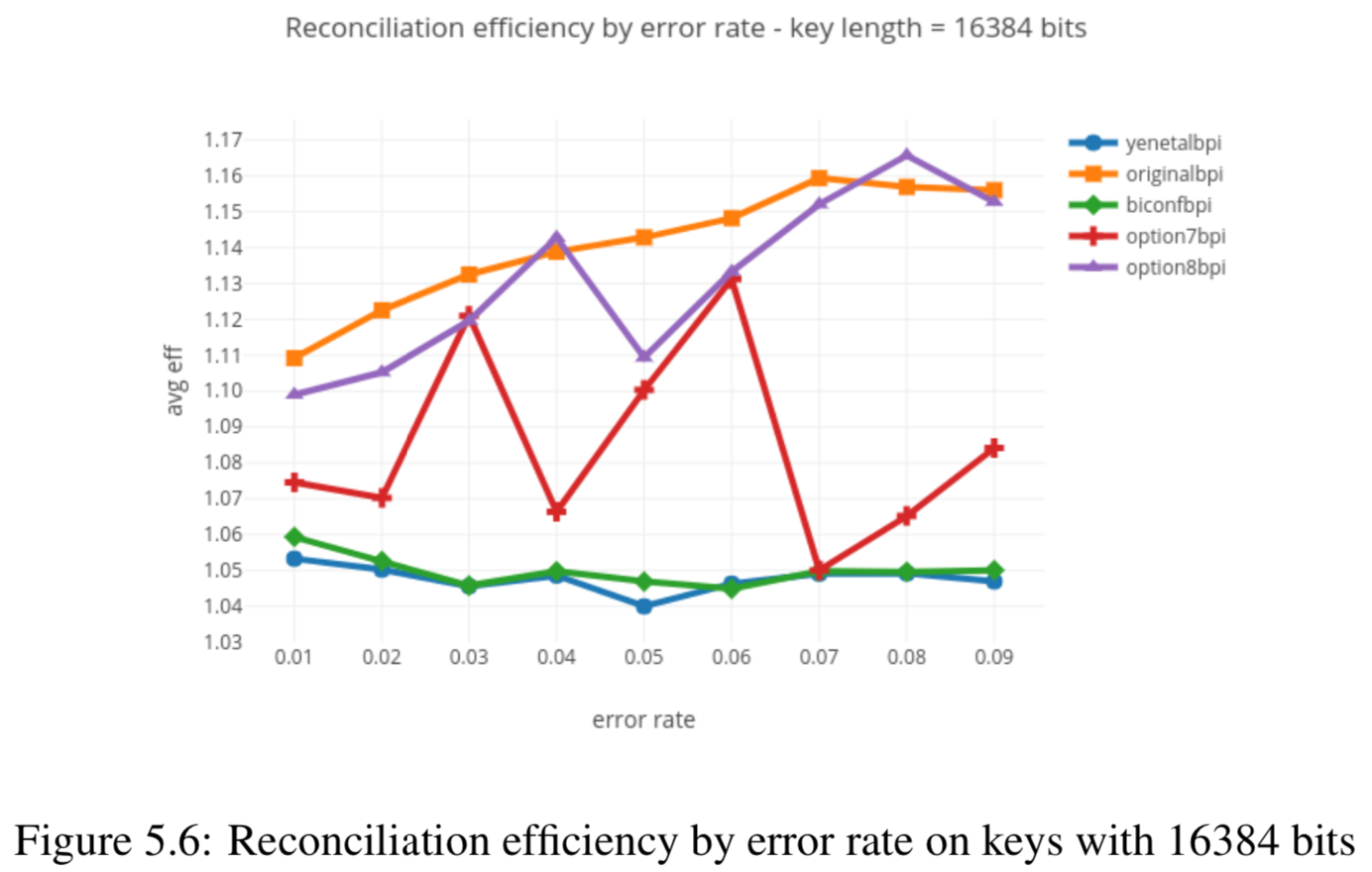
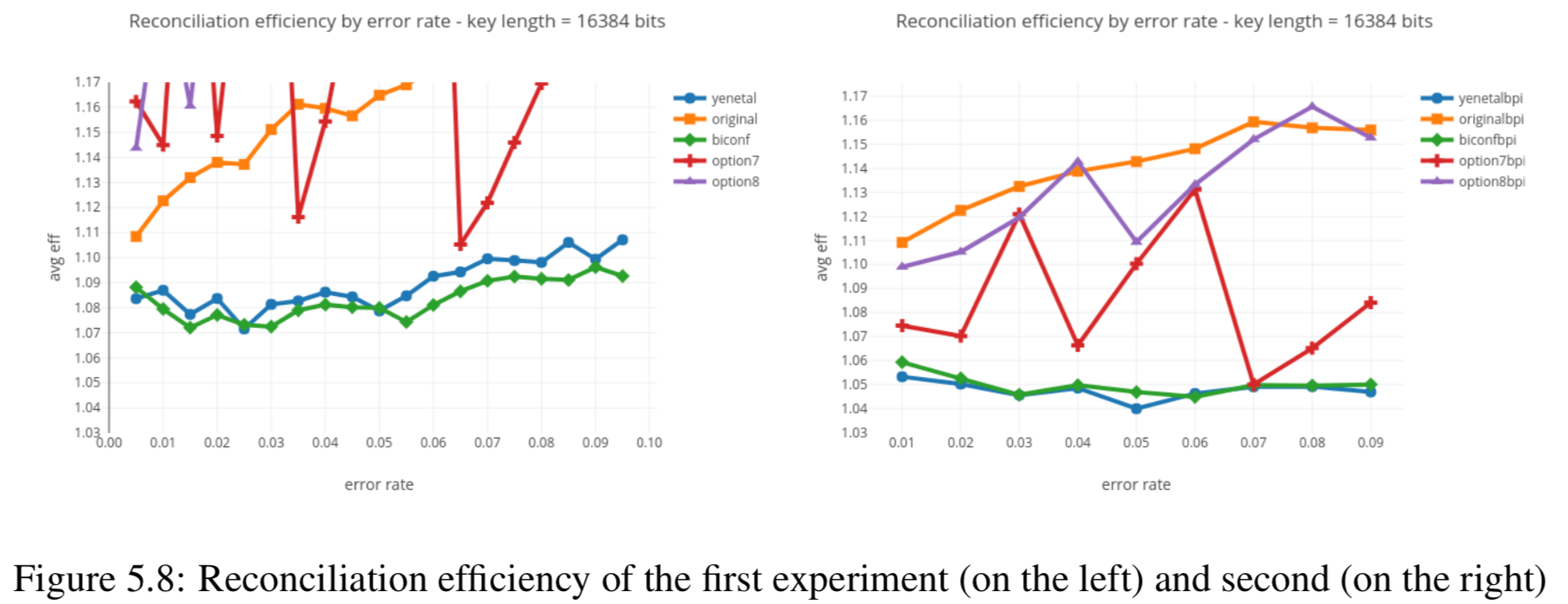
Results reported in thesis:
Results produced by Cascade-CPP:
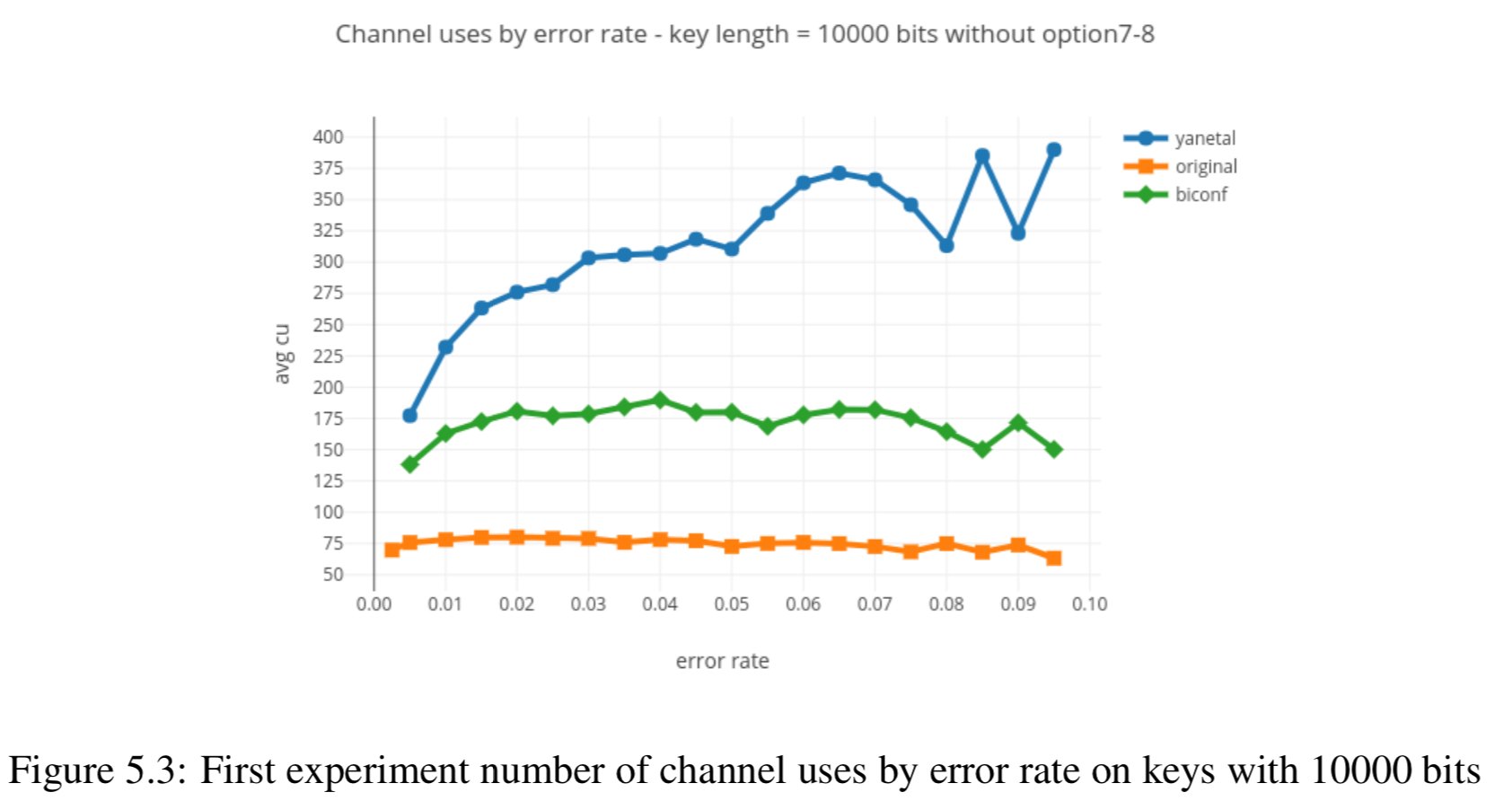
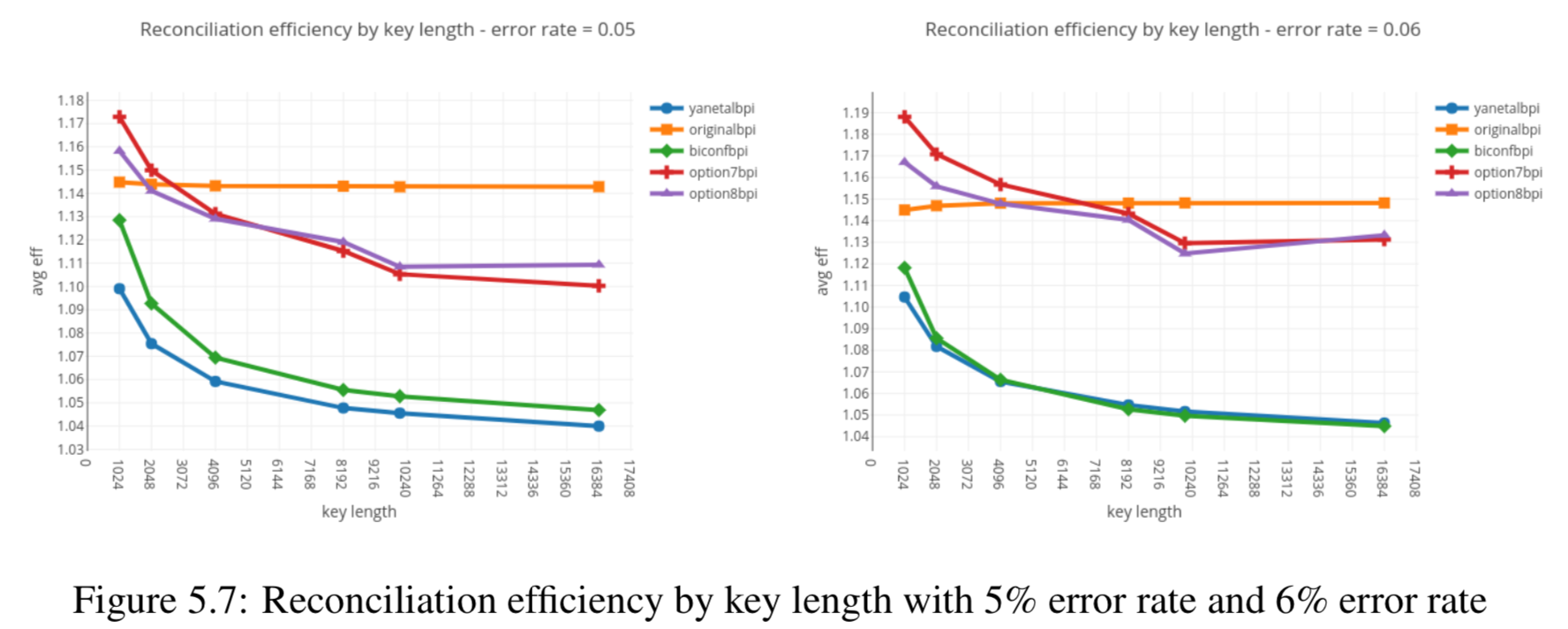
Results reported in thesis:
Results produced by Cascade-CPP:
Results reported in thesis:
Results not produced by Cascade-CPP.
Results reported in thesis:
Results produced by Cascade-CPP:
Results reported in thesis:
Results not yet produced by Cascade-CPP.
Results reported in thesis:
Results not yet produced by Cascade-CPP.
Results reported in thesis:
Results not yet produced by Cascade-CPP.
Results reported in thesis:
Results not yet produced by Cascade-CPP.
Results reported in thesis:
Results not yet produced by Cascade-CPP.