Arraymancer - A n-dimensional tensor (ndarray) library.
Arraymancer is a tensor (N-dimensional array) project in Nim. The main focus is providing a fast and ergonomic CPU, Cuda and OpenCL ndarray library on which to build a scientific computing and in particular a deep learning ecosystem.
The library is inspired by Numpy and PyTorch. The library provides ergonomics very similar to Numpy, Julia and Matlab but is fully parallel and significantly faster than those libraries. It is also faster than C-based Torch.
Note: While Nim is compiled and does not offer an interactive REPL yet (like Jupyter), it allows much faster prototyping than C++ due to extremely fast compilation times. Arraymancer compiles in about 5 seconds on my dual-core MacBook.
Show me some code
Arraymancer tutorial is available here.
Here is a preview of Arraymancer syntax.
Tensor creation and slicing
import math, arraymancer, future
const
x = @[1, 2, 3, 4, 5]
y = @[1, 2, 3, 4, 5]
var
vandermonde: seq[seq[int]]
row: seq[int]
vandermonde = newSeq[seq[int]]()
for i, xx in x:
row = newSeq[int]()
vandermonde.add(row)
for j, yy in y:
vandermonde[i].add(xx^yy)
let foo = vandermonde.toTensor()
echo foo
# Tensor of shape 5x5 of type "int" on backend "Cpu"
# |1 1 1 1 1|
# |2 4 8 16 32|
# |3 9 27 81 243|
# |4 16 64 256 1024|
# |5 25 125 625 3125|
echo foo[1..2, 3..4] # slice
# Tensor of shape 2x2 of type "int" on backend "Cpu"
# |16 32|
# |81 243|Reshaping and concatenation
import arraymancer, sequtils
let a = toSeq(1..4).toTensor.reshape(2,2)
let b = toSeq(5..8).toTensor.reshape(2,2)
let c = toSeq(11..16).toTensor
let c0 = c.reshape(3,2)
let c1 = c.reshape(2,3)
echo concat(a,b,c0, axis = 0)
# Tensor of shape 7x2 of type "int" on backend "Cpu"
# |1 2|
# |3 4|
# |5 6|
# |7 8|
# |11 12|
# |13 14|
# |15 16|
echo concat(a,b,c1, axis = 1)
# Tensor of shape 2x7 of type "int" on backend "Cpu"
# |1 2 5 6 11 12 13|
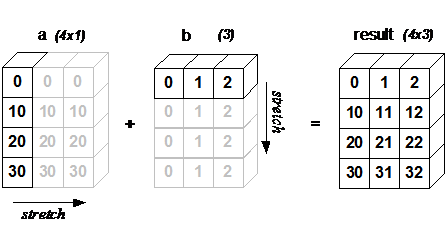
# |3 4 7 8 14 15 16|Broadcasting
Image from Scipy
import arraymancer
let j = [0, 10, 20, 30].toTensor.reshape(4,1)
let k = [0, 1, 2].toTensor.reshape(1,3)
echo j .+ k
# Tensor of shape 4x3 of type "int" on backend "Cpu"
# |0 1 2|
# |10 11 12|
# |20 21 22|
# |30 31 32|Table of Contents
- Arraymancer - A n-dimensional tensor (ndarray) library.
3 reasons why Arraymancer
The Python community is struggling to bring Numpy up-to-speed
- Numba JIT compiler
- Dask delayed parallel computation graph
- Cython to ease numerical computations in Python
- Due to the GIL shared-memory parallelism (OpenMP) is not possible in pure Python
- Use "vectorized operations" (i.e. don't use for loops in Python)
Why not use in a single language with all the blocks to build the most efficient scientific computing library with Python ergonomics.
OpenMP batteries included.
A researcher workflow is a fight against inefficiencies
Researchers in a heavy scientific computing domain often have the following workflow: Mathematica/Matlab/Python/R (prototyping) -> C/C++/Fortran (speed, memory)
Why not use in a language as productive as Python and as fast as C? Code once, and don't spend months redoing the same thing at a lower level.
Bridging the gap between deep learning research and production
The deep learning frameworks are currently in two camps:
- Research: Theano, Tensorflow, Keras, Torch, PyTorch
- Production: Caffe, Darknet, (Tensorflow)
Furthermore, Python preprocessing steps, unless using OpenCV, often needs a custom implementation (think text/speech preprocessing on phones).
- Managing and deploying Python (2.7, 3.5, 3.6) and packages version in a robust manner requires devops-fu (virtualenv, Docker, ...)
- Python data science ecosystem does not run on embedded devices (Nvidia Tegra/drones) or mobile phones, especially preprocessing dependencies.
- Tensorflow is supposed to bridge the gap between research and production but its syntax and ergonomics are a pain to work with. Like for researchers, you need to code twice, "Prototype in Keras, and when you need low-level --> Tensorflow".
- Deployed models are static, there is no interface to add a new observation/training sample to any framework, what if you want to use a model as a webservice with online learning?
So why Arraymancer ?
All those pain points may seem like a huge undertaking however thanks to the Nim language, we can have Arraymancer:
- Be as fast as C
- Accelerated routines with Intel MKL/OpenBLAS or even NNPACK
- Access to CUDA and CuDNN and generate custom CUDA kernels on the fly via metaprogramming.
- A Python-like syntax with custom operators
a * bfor tensor multiplication instead ofa.dot(b)(Numpy/Tensorflow) ora.mm(b)(Torch) - Numpy-like slicing ergonomics
t[0..4, 2..10|2] - For everything that Nim doesn't have yet, you can use Nim bindings to C, C++, Objective-C or Javascript to bring it to Nim. Nim also has unofficial Python->Nim and Nim->Python wrappers.
Future ambitions
Because apparently to be successful you need a vision, I would like Arraymancer to be:
- The go-to tool for Deep Learning video processing. I.e.
vid = load_video("./cats/youtube_cat_video.mkv") - Target javascript, WebAssembly, Apple Metal, ARM devices, AMD Rocm, OpenCL, you name it.
- The base of a Starcraft II AI bot.
- Target cryptominers FPGAs because they drove the price of GPUs for honest deep-learners too high.
Installation
Nim is available in some Linux repositories and on Homebrew for macOS.
I however recommend installing Nim in your user profile via choosenim. Once choosenim installed Nim, you can nimble install arraymancer which will pull the latest arraymancer release and all its dependencies.
To install Arraymancer development version you can use nimble install arraymancer@#head.
Full documentation
Detailed API is available on Arraymancer official documentation.
Features
For now Arraymancer is mostly at the ndarray stage, a vision package and a deep learning demo are available with logistic regression and perceptron from scratch.
You can also check the detailed example or benchmark perceptron for a preview of Arraymancer deep learning usage.
Available autograd and neural networks features are detailed in the technical reference part of the documentation.
Arraymancer as a Deep Learning library
Note: The final interface is still work in progress.
Handwritten digit recognition with Arraymancer
From example 2.
import src/arraymancer, random
# This is an early minimum viable example of handwritten digits recognition.
# It uses convolutional neural networks to achieve high accuracy.
#
# Data files (MNIST) can be downloaded here http://yann.lecun.com/exdb/mnist/
# and must be decompressed in "./build/" (or change the path "build/..." below)
#
# Note:
# In the future, model, weights and optimizer definition will be streamlined.
# Make the results reproducible by initializing a random seed
randomize(42)
let
ctx = newContext Tensor[float32] # Autograd/neural network graph
n = 32 # Batch size
let
# Training data is 60k 28x28 greyscale images from 0-255,
# neural net prefers input rescaled to [0, 1] or [-1, 1]
x_train = read_mnist_images("build/train-images.idx3-ubyte").astype(float32) / 255'f32
# Change shape from [N, H, W] to [N, C, H, W], with C = 1 (unsqueeze). Convolution expect 4d tensors
# And store in the context to track operations applied and build a NN graph
X_train = ctx.variable x_train.unsqueeze(1)
# Labels are uint8, we must convert them to int
y_train = read_mnist_labels("build/train-labels.idx1-ubyte").astype(int)
# Idem for testing data (10000 images)
x_test = read_mnist_images("build/t10k-images.idx3-ubyte").astype(float32) / 255'f32
X_test = ctx.variable x_test.unsqueeze(1)
y_test = read_mnist_labels("build/t10k-labels.idx1-ubyte").astype(int)
# Config (API is not finished)
let
# We randomly initialize all weights and bias between [-0.5, 0.5]
# In the future requires_grad will be automatically set for neural network layers
cv1_w = ctx.variable(
randomTensor(20, 1, 5, 5, 1'f32) .- 0.5'f32, # Weight of 1st convolution
requires_grad = true
)
cv1_b = ctx.variable(
randomTensor(20, 1, 1, 1'f32) .- 0.5'f32, # Bias of 1st convolution
requires_grad = true
)
cv2_w = ctx.variable(
randomTensor(50, 20, 5, 5, 1'f32) .- 0.5'f32, # Weight of 2nd convolution
requires_grad = true
)
cv2_b = ctx.variable(
randomTensor(50, 1, 1, 1'f32) .- 0.5'f32, # Bias of 2nd convolution
requires_grad = true
)
fc3 = ctx.variable(
randomTensor(500, 800, 1'f32) .- 0.5'f32, # Fully connected: 800 in, 500 ou
requires_grad = true
)
classifier = ctx.variable(
randomTensor(10, 500, 1'f32) .- 0.5'f32, # Fully connected: 500 in, 10 classes out
requires_grad = true
)
proc model[TT](x: Variable[TT]): Variable[TT] =
# The formula of the output size of convolutions and maxpools is:
# H_out = (H_in + (2*padding.height) - kernel.height) / stride.height + 1
# W_out = (W_in + (2*padding.width) - kernel.width) / stride.width + 1
let cv1 = x.conv2d(cv1_w, cv1_b).relu() # Conv1: [N, 1, 28, 28] --> [N, 20, 24, 24] (kernel: 5, padding: 0, strides: 1)
let mp1 = cv1.maxpool2D((2,2), (0,0), (2,2)) # Maxpool1: [N, 20, 24, 24] --> [N, 20, 12, 12] (kernel: 2, padding: 0, strides: 2)
let cv2 = mp1.conv2d(cv2_w, cv2_b).relu() # Conv2: [N, 20, 12, 12] --> [N, 50, 8, 8] (kernel: 5, padding: 0, strides: 1)
let mp2 = cv2.maxpool2D((2,2), (0,0), (2,2)) # Maxpool1: [N, 50, 8, 8] --> [N, 50, 4, 4] (kernel: 2, padding: 0, strides: 2)
let f = mp2.flatten # [N, 50, 4, 4] -> [N, 800]
let hidden = f.linear(fc3).relu # [N, 800] -> [N, 500]
result = hidden.linear(classifier) # [N, 500] -> [N, 10]
# Stochastic Gradient Descent (API will change)
let optim = newSGD[float32](
cv1_w, cv1_b, cv2_w, cv2_b, fc3, classifier, 0.01f # 0.01 is the learning rate
)
# Learning loop
for epoch in 0 ..< 5:
for batch_id in 0 ..< X_train.value.shape[0] div n: # some at the end may be missing, oh well ...
# minibatch offset in the Tensor
let offset = batch_id * n
let x = X_train[offset ..< offset + n, _]
let target = y_train[offset ..< offset + n]
# Running through the network and computing loss
let clf = x.model
let loss = clf.sparse_softmax_cross_entropy(target)
if batch_id mod 200 == 0:
# Print status every 200 batches
echo "Epoch is: " & $epoch
echo "Batch id: " & $batch_id
echo "Loss is: " & $loss.value.data[0]
# Compute the gradient (i.e. contribution of each parameter to the loss)
loss.backprop()
# Correct the weights now that we have the gradient information
optim.update()
# Validation (checking the accuracy/generalization of our model on unseen data)
ctx.no_grad_mode:
echo "\nEpoch #" & $epoch & " done. Testing accuracy"
# To avoid using too much memory we will compute accuracy in 10 batches of 1000 images
# instead of loading 10 000 images at once
var score = 0.0
var loss = 0.0
for i in 0 ..< 10:
let y_pred = X_test[i ..< i+1000, _].model.value.softmax.argmax(axis = 1).indices.squeeze
score += accuracy_score(y_test[i ..< i+1000, _], y_pred)
loss += X_test[i ..< i+1000, _].model.sparse_softmax_cross_entropy(y_test[i ..< i+1000, _]).value.data[0]
score /= 10
loss /= 10
echo "Accuracy: " & $(score * 100) & "%"
echo "Loss: " & $loss
echo "\n"
############# Output ############
# Epoch is: 0
# Batch id: 0
# Loss is: 132.9124755859375
# Epoch is: 0
# Batch id: 200
# Loss is: 2.301989078521729
# Epoch is: 0
# Batch id: 400
# Loss is: 1.155071973800659
# Epoch is: 0
# Batch id: 600
# Loss is: 1.043337464332581
# Epoch is: 0
# Batch id: 800
# Loss is: 0.58299720287323
# Epoch is: 0
# Batch id: 1000
# Loss is: 0.5417937040328979
# Epoch is: 0
# Batch id: 1200
# Loss is: 0.6955615282058716
# Epoch is: 0
# Batch id: 1400
# Loss is: 0.4742314517498016
# Epoch is: 0
# Batch id: 1600
# Loss is: 0.3307125866413116
# Epoch is: 0
# Batch id: 1800
# Loss is: 0.6455222368240356
# Epoch #0 done. Testing accuracy
# Accuracy: 83.24999999999999%
# Loss: 0.5828457295894622
# Epoch is: 1
# Batch id: 0
# Loss is: 0.5344035029411316
# Epoch is: 1
# Batch id: 200
# Loss is: 0.4455387890338898
# Epoch is: 1
# Batch id: 400
# Loss is: 0.1642555445432663
# Epoch is: 1
# Batch id: 600
# Loss is: 0.5191419124603271
# Epoch is: 1
# Batch id: 800
# Loss is: 0.2091695368289948
# Epoch is: 1
# Batch id: 1000
# Loss is: 0.2661008834838867
# Epoch is: 1
# Batch id: 1200
# Loss is: 0.405451238155365
# Epoch is: 1
# Batch id: 1400
# Loss is: 0.1397259384393692
# Epoch is: 1
# Batch id: 1600
# Loss is: 0.526863694190979
# Epoch is: 1
# Batch id: 1800
# Loss is: 0.5916416645050049
# Epoch #1 done. Testing accuracy
# Accuracy: 88.49000000000001%
# Loss: 0.3582650691270828Tensors on CPU and on Cuda
Tensors and CudaTensors do not have the same features implemented yet. Also Cuda Tensors can only be float32 or float64 while Cpu Tensor can be integers, string, boolean or any custom object.
Here is a comparative table, not that this feature set is developing very rapidly.
| Action | Tensor | CudaTensor |
|---|---|---|
| Accessing tensor properties | [x] | [x] |
| Tensor creation | [x] | by converting a cpu Tensor |
| Accessing or modifying a single value | [x] | [] |
| Iterating on a Tensor | [x] | [] |
| Slicing a Tensor | [x] | [x] |
Slice mutation a[1,_] = 10 |
[x] | [] |
Comparison == |
[x] | Coming soon |
| Element-wise basic operations | [x] | [x] |
| Universal functions | [x] | [x] |
| Automatically broadcasted operations | [x] | Coming soon |
| Matrix-Matrix and Matrix-Vector multiplication | [x] | [x] Note that sliced CudaTensors must explicitly be made contiguous for the moment |
| Displaying a tensor | [x] | [x] |
| Higher-order functions (map, apply, reduce, fold) | [x] | Apply, but only internally |
| Transposing | [x] | [x] |
| Converting to contiguous | [x] | [x] |
| Reshaping | [x] | [] |
| Explicit broadcast | [x] | Coming soon |
| Permuting dimensions | [x] | Coming soon |
| Concatenating tensors along existing dimension | [x] | [] |
| Squeezing singleton dimension | [x] | Coming soon |
| Slicing + squeezing | [x] | Coming soon |
Speed
Arraymancer is fast, how it achieves its speed under the hood is detailed here.
Micro benchmark: Int64 matrix multiplication
Integers seem to be the abandoned children of ndarrays and tensors libraries. Everyone is optimising the hell of floating points. Not so with Arraymancer:
Archlinux, E3-1230v5 (Skylake quad-core 3.4 GHz, turbo 3.8)
Input 1500x1500 random large int64 matrix
Arraymancer 0.2.90 (master branch 2017-10-10)
| Language | Speed | Memory |
|---|---|---|
| Nim 0.17.3 (devel) + OpenMP | 0.36s | 55.5 MB |
| Julia v0.6.0 | 3.11s | 207.6 MB |
| Python 3.6.2 + Numpy 1.12 compiled from source | 8.03s | 58.9 MB |
MacOS + i5-5257U (Broadwell dual-core mobile 2.7GHz, turbo 3.1)
Input 1500x1500 random large int64 matrix
Arraymancer 0.2.90 (master branch 2017-10-31)
no OpenMP compilation: nim c -d:native -d:release --out:build/integer_matmul --nimcache:./nimcache benchmarks/integer_matmul.nim
with OpenMP: nim c -d:openmp --cc:gcc --gcc.exe:"/usr/local/bin/gcc-6" --gcc.linkerexe:"/usr/local/bin/gcc-6" -d:native -d:release --out:build/integer_matmul --nimcache:./nimcache benchmarks/integer_matmul.nim
| Language | Speed | Memory |
|---|---|---|
| Nim 0.18.0 (devel) - GCC 6 + OpenMP | 0.95s | 71.9 MB |
| Nim 0.18.0 (devel) - Apple Clang 9 - no OpenMP | 1.73s | 71.7 MB |
| Julia v0.6.0 | 4.49s | 185.2 MB |
| Python 3.5.2 + Numpy 1.12 | 9.49s | 55.8 MB |
Benchmark setup is in the ./benchmarks folder and similar to (stolen from) Kostya's. Note: Arraymancer float matmul is as fast as Julia Native Thread.
Logistic regression
On the demo benchmark, Arraymancer is faster than Torch in v0.2.90.
CPU
| Framework | Backend | Forward+Backward Pass Time |
|---|---|---|
| Arraymancer v0.2.90 | OpenMP + MKL | 0.458ms |
| Torch7 | MKL | 0.686ms |
| Numpy | MKL | 0.723ms |
GPU
| Framework | Backend | Forward+Backward Pass Time |
|---|---|---|
| Arraymancer v0.2.90 | Cuda | WIP |
| Torch7 | Cuda | 0.286ms |
DNN - 3 hidden layers
CPU
| Framework | Backend | Forward+Backward Pass Time |
|---|---|---|
| Arraymancer v0.2.90 | OpenMP + MKL | 2.907ms |
| PyTorch | MKL | 6.797ms |
GPU
| Framework | Backend | Forward+Backward Pass Time |
|---|---|---|
| Arraymancer v0.2.90 | Cuda | WIP |
| PyTorch | Cuda | 4.765ms |
Intel(R) Core(TM) i7-3770K CPU @ 3.50GHz, gcc 7.2.0, MKL 2017.17.0.4.4, OpenBLAS 0.2.20, Cuda 8.0.61, Geforce GTX 1080 Ti, Nim 0.18.0
In the future, Arraymancer will leverage Nim compiler to automatically fuse operations
like alpha A*B + beta C or a combination of element-wise operations. This is already done to fuse toTensor and reshape.