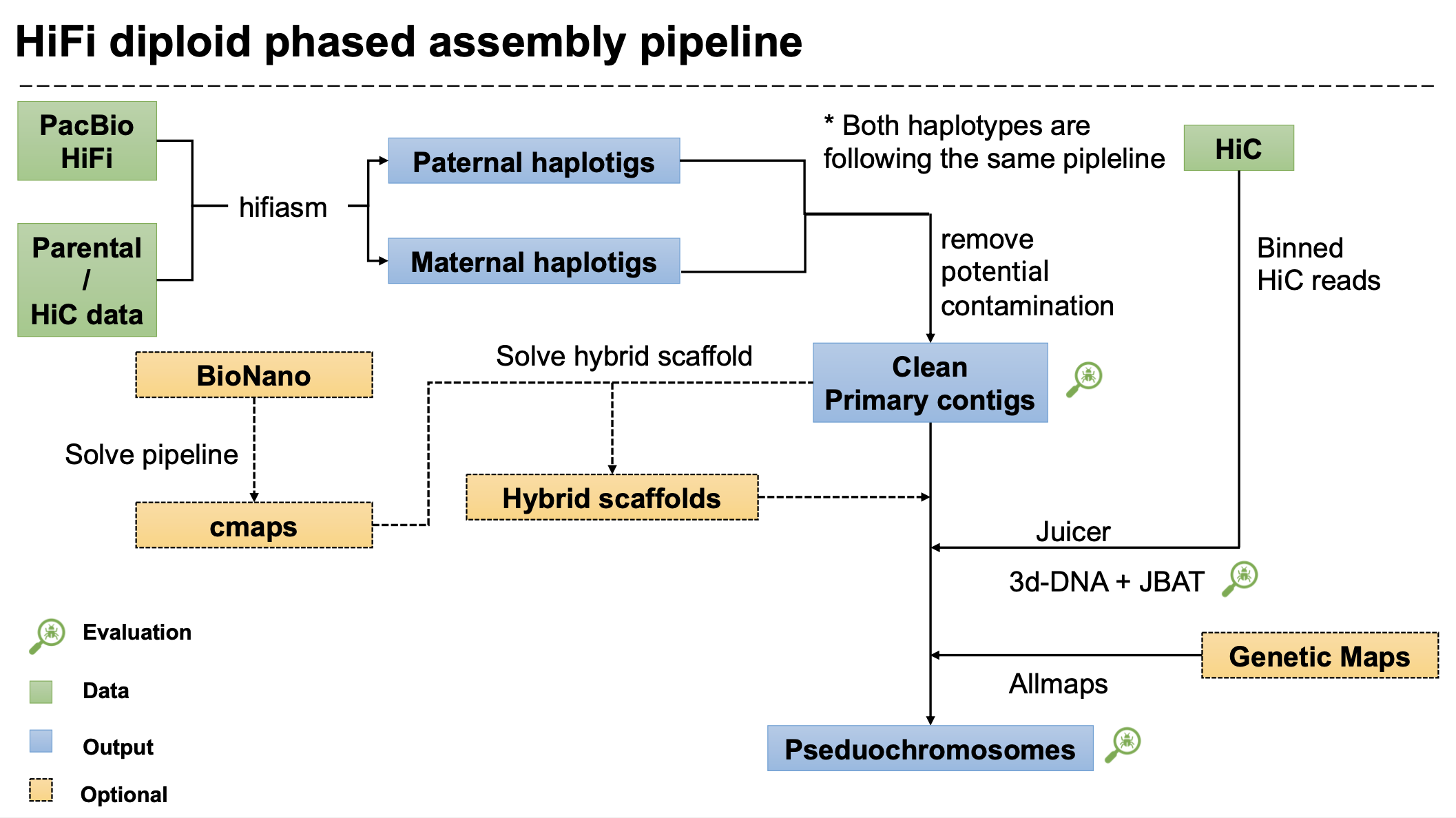
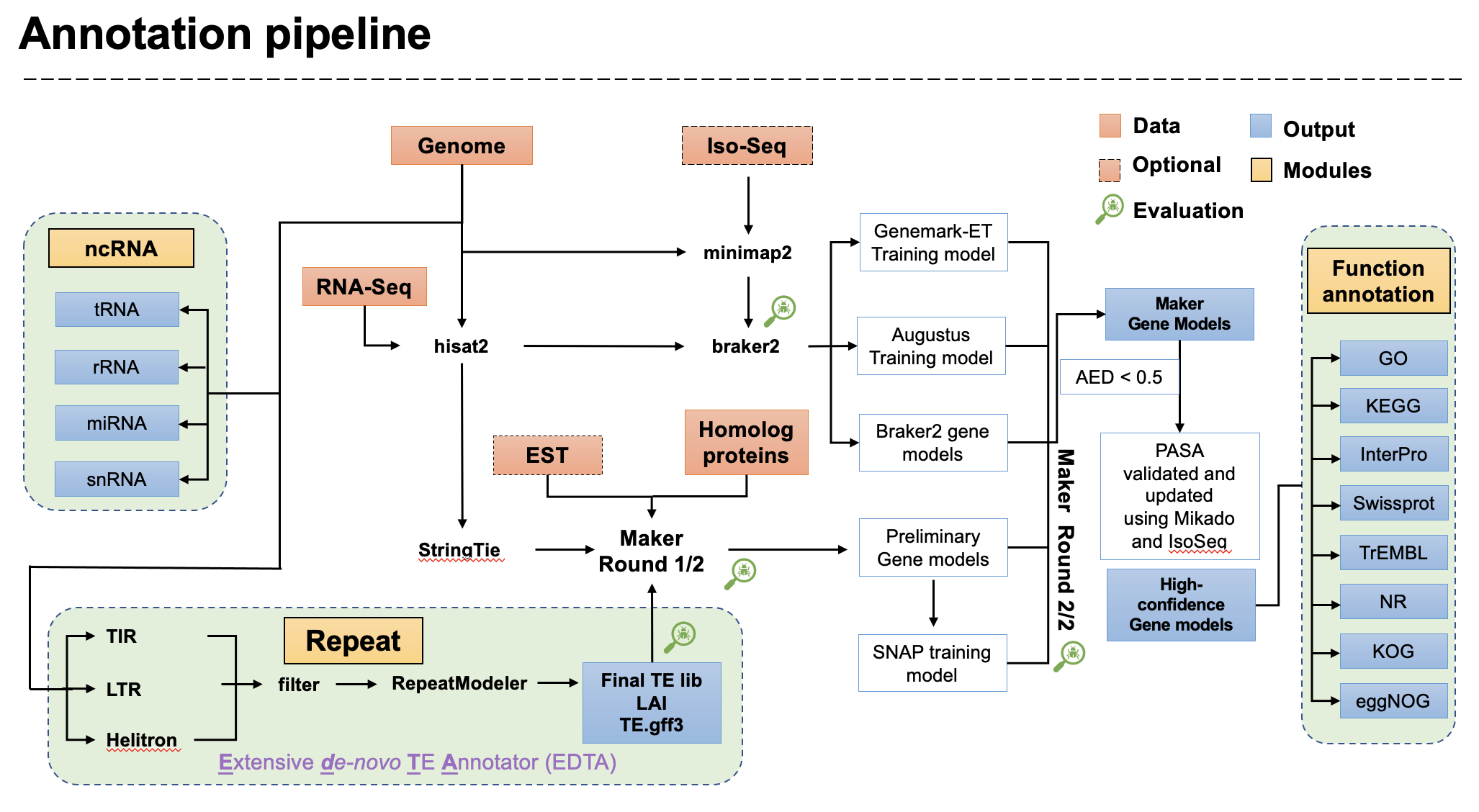
This Snakemake pipeline aims for plant genome assembly with HiFi data and genome annotation with RNA-Seq/IsoSeq.
Modified from the VGP assembly VGP tutorial
- Maker running inside
snakemakestill have the problem since the NSF lock and submit system is already included. - IsoSeq integration is still in development. For diploid or polyploidy, FLNC mapping is better than the clustering of the PacBio pipeline if you have haplotype-resolved assembly.
- Automatic pipeline always confused the complex gene clusters (metabolic gene clusters / NLR gene clusters), please manually curation it before any further interpretation.
WebApolloorIGV-GSAmancould be a good start for manual curation. (https://www.bilibili.com/video/BV1x84y1z7ZW/)